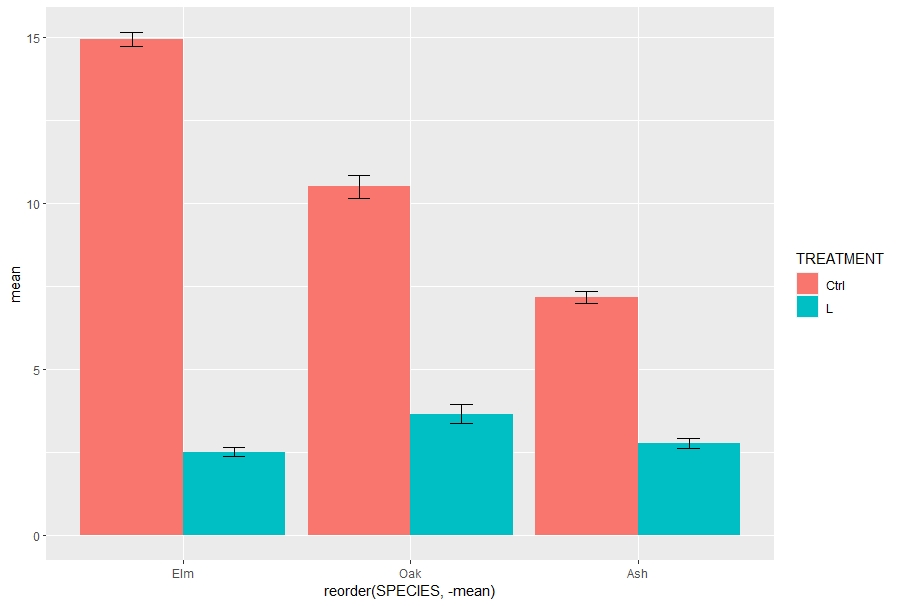
I have some data that shows a treatment effect for a range of tree species, and I'm performing a one-way anova of RESULT ~ TREATMENT for each species. Using dplyr, I've created a new data frame of treatment means grouped by species, as well as standard errors and deviations. I'd like to add the compact letter display results to that same data frame.
Here's some dummy data to play with
library(dplyr)
library(tidyverse)
library(ggplot2)
# Generate species
species <- rep (c("Oak", "Elm", "Ash"), each = 10)
# Generate treatments
dose_1 <- rep (c("Ctrl"), 30)
dose_2 <- rep (c ("L"), 30)
# Generate results
result_1 <- c((runif(10, 9, 12)), runif(10, 14, 16), runif(10, 6, 8), (runif(10, 2, 5)), runif(10, 1, 4), runif(10, 2, 4))
# Combine into a sinlge dataframe
data <- data.frame (SPECIES = rep(species, 2), TREATMENT = c(dose_1, dose_2), RESULT = result_1)
# Consolidate into a new data frame for ggplot and add errors
dat <- data %>%
group_by(SPECIES, TREATMENT) %>%
summarise(
n=n(),
mean=mean(RESULT),
sd=sd(RESULT)
) %>%
mutate( se=sd/sqrt(n)) %>%
mutate( ic=se * qt((1-0.05)/2 .5, n-1))
# Plot the results
ggplot(dat, aes(x= reorder(SPECIES, -mean), y = mean, fill = TREATMENT))
geom_bar(position = 'dodge', stat = 'identity')
geom_errorbar(aes(ymin = mean-se, ymax = mean se), position = position_dodge(.9), width = 0.2)
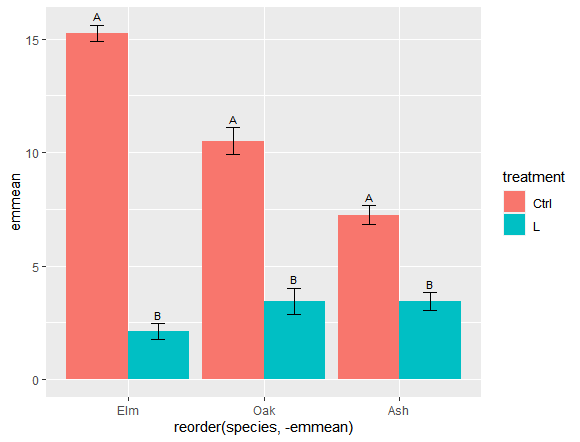
Now I'd like to get the compact letter display results from a Tukey test for the treatment effect on each species and add it to dat
# First perform anova tests for treatment effects for each species
df_aov <- data %>%
dplyr::group_by(SPECIES) %>%
tidyr::nest() %>%
dplyr::mutate(.data = .,
aov_results = data %>% purrr::map(.x = ., .f = ~ summary(aov(RESULT ~ TREATMENT, data = .))))
# Inspect the results
df_aov$aov_results[[1]]
df_aov$aov_results[[2]]
df_aov$aov_results[[3]]
At this point I can only perform the Tukey test on the anova results for each species, like this
# Tukey's test
tukey <- TukeyHSD(df_aov$aov_results[[1]])
# compact letter display
cld <- multcompLetters4(df_aov$aov_results[[1]], tukey)
What I'd like to be able to do is perform the Tukey tests as batch on all the species and add the compact letter display results to the dat data frame that I'm using for ggplot. dat should end up looking something like this at the end
dat
# A tibble: 6 x 8
# Groups: SPECIES [3]
SPECIES TREATMENT n mean sd se ic cld
<chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <chr>
1 Ash Ctrl 10 7.17 0.556 0.176 0.398 a
2 Ash L 10 2.78 0.454 0.143 0.324 b
3 Elm Ctrl 10 15.0 0.653 0.206 0.467 a
4 Elm L 10 2.52 0.468 0.148 0.335 b
5 Oak Ctrl 10 10.5 1.13 0.357 0.808 a
6 Oak L 10 3.66 0.895 0.283 0.640 b
I'd then like to add a line of code to the ggplot like this, so I can add the compact letter display results to each species above the error bars.
[first part of the plot code]
geom_text(aes(label = cld, y = mean se), vjust = -0.5, position = position_dodge(0.9),size = 3)
CodePudding user response:
starting with dataframe data of your example, you could do the following:
- create dataframe with descriptive stats (as in your example)
library(multcompView)
library(dplyr)
df_summary <-
data %>%
group_by(SPECIES, TREATMENT) %>%
summarise(n = n(),
sd = sd(RESULT, na.rm = TRUE),
mean=mean(RESULT)
) %>%
mutate(se = sd/sqrt(n),
ic = se * qt((1-0.05)/2 .5, n-1)
)
- create dataframe with Tukey stats (note the use of
rowwise):
df_tukey <-
data %>%
group_by(SPECIES) %>%
nest %>%
rowwise %>% ## !important
mutate(aov_result = list(aov(RESULT ~ TREATMENT, data = data)),
tukey = list(TukeyHSD(aov_result)),
cld = list(multcompLetters4(aov_result, tukey)$TREATMENT$Letters )
) %>%
unnest(cld) %>%
select(SPECIES, LETTER = cld) %>%
mutate(TREATMENT = names(LETTER))
- join both:
df_summary %>%
left_join(df_tukey)
CodePudding user response: