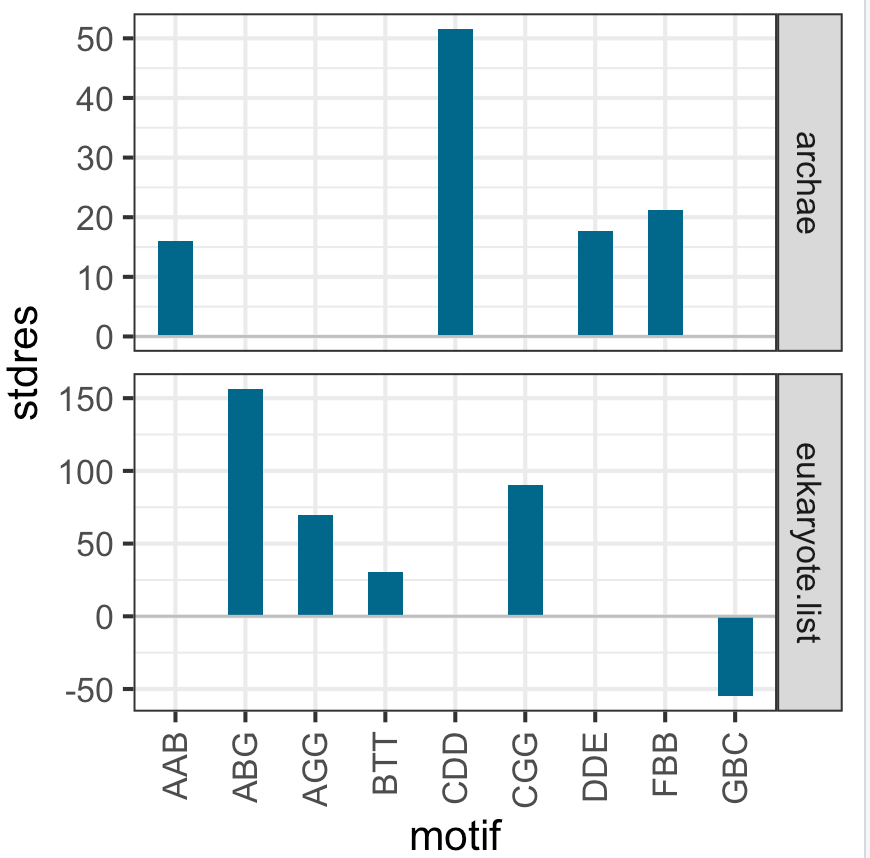
I have a list of data frames,
>head(df.list.xyg[["archae.list"]])
motif obs pred prop stat pval stdres
AAB 1189 760.1757 0.05556028 11811.94 0 16.00425
CDD 1058 249.7147 0.01825133 11811.94 0 51.62291
DDE 771 415.1314 0.03034143 11811.94 0 17.73730
FBB 544 226.3529 0.01654385 11811.94 0 21.28994
>head(df.list.xyg[["eukaryote.list"]])
motif obs pred prop stat pval stdres
ABG 82015 48922.33 0.08773749 321891.7 0 156.64562
GBC 51601 64768.42 0.11615591 321891.7 0 -55.03402
AGG 41922 30136.56 0.05404701 321891.7 0 69.80141
CGG 25545 14757.24 0.02646569 321891.7 0 90.00215
BTT 15795 12433.58 0.02229843 321891.7 0 30.48747
I would like to
- barplot motif versus stdres such that the plots are displayed in single column but two rows, and
- label each plot corresponding to part of the file name "archae", eukaryote and so on.
After searching around, the following code snippet does the job but only partly. It only plots the last dataset. I assume it is "overwriting" the earlier dataset. How do I fix this? Can this be achieved by face_wrap or facet_grid?
myplots<-lapply(df.list.xyg,
function(x)
p<-ggplot(x,aes(x=motif,y=stdres))
geom_bar(stat="identity",width=0.5, color="blue", fill="gray")
)
print (myplots)
CodePudding user response:
If you just want the plots to appear together in a single plotting window, you can use facets. You can bind the rows of your data frames together with dplyr::bind_rows, which will create an id column to label which data frame each row belongs to. We facet by this ID variable.
library(ggplot2)
ggplot(dplyr::bind_rows(df.list.xyg, .id = "Kingdom"), aes(motif, stdres))
geom_col(width = 0.5, fill = "deepskyblue4")
geom_hline(yintercept = 0, color = "gray75")
facet_grid(.~Kingdom)
theme_bw(base_size = 16)
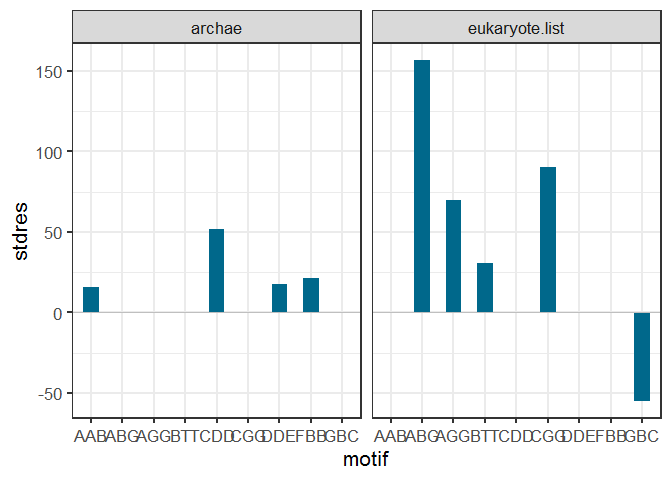
If you have lots of different data frames in your list, facet_grid may be better than facet_wrap.
Depending on how you wish to present the data, you may prefer to save individual plots to files using ggsave inside your lapply
Reproducible data taken from question
df.list.xyg <- list(archae = structure(list(motif = c("AAB",
"CDD", "DDE", "FBB"), obs = c(1189L, 1058L, 771L, 544L),
pred = c(760.1757, 249.7147,
415.1314, 226.3529), prop = c(0.05556028, 0.01825133, 0.03034143,
0.01654385), stat = c(11811.94, 11811.94, 11811.94, 11811.94),
pval = c(0L, 0L, 0L, 0L), stdres = c(16.00425, 51.62291,
17.7373, 21.28994)), class = "data.frame", row.names = c(NA,
-4L)), eukaryote.list = structure(list(motif = c("ABG", "GBC",
"AGG", "CGG", "BTT"), obs = c(82015L, 51601L, 41922L, 25545L,
15795L), pred = c(48922.33, 64768.42, 30136.56, 14757.24, 12433.58
), prop = c(0.08773749, 0.11615591, 0.05404701, 0.02646569, 0.02229843
), stat = c(321891.7, 321891.7, 321891.7, 321891.7, 321891.7),
pval = c(0L, 0L, 0L, 0L, 0L), stdres = c(156.64562, -55.03402,
69.80141, 90.00215, 30.48747)), class = "data.frame", row.names = c(NA,
-5L)))