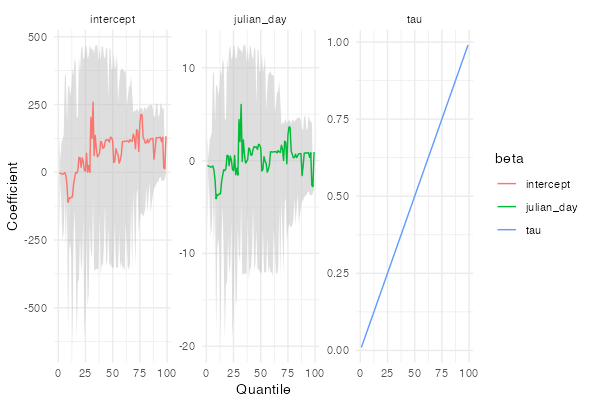
Thank you very much for the help, if I am missing to give some informations do not hesitate to ask, I will edit my post.
CodePudding user response:
As mentioned in the comments, you can do this with geom_ribbon(), you just need to merge the CI data with the model coefficient data.
CIs_to_plot <- map_dfr(1:dim(CI_mod1)[3], ~as_tibble(CI_mod1[,,.x], rownames = "pctle"),
.id = "Quantile") %>%
pivot_wider(names_from = "pctle", values_from = c("V1", "V2")) %>%
rename("intercept.low" = `V1_2.5%`, "intercept.high" = `V1_97.5%`,
"julian_day.low" =`V2_2.5%`, "julian_day.high" = `V2_97.5%`) %>%
pivot_longer(-Quantile, names_sep = "\\.", names_to = c("beta", ".value")) %>%
mutate(Quantile = parse_number(Quantile))
model_to_plot <- model1 %>%
mutate(Quantile=row_number()) %>%
pivot_longer(!Quantile,names_to="beta",values_to = "Coefficient")
model_to_plot %>%
left_join(CIs_to_plot, by = c("Quantile", "beta")) %>%
ggplot(aes(Quantile,Coefficient))
geom_ribbon(aes(ymin = low, ymax = high), fill = "grey", alpha = .5)
geom_line(aes(color=beta))
facet_wrap(~beta, scales="free_y")
theme_minimal()