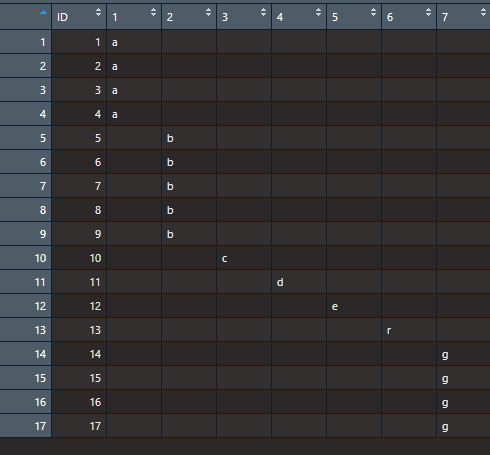
I am after reshaping the data, and get the below, see screenshot. How can I push the values up? Such that column 2 b row 5, value b, will go into column 2, element 1? Like removing the ID vector and pushing the remaining data upwwards.
I have tried d <- apply(d, 2, sort) and it does not work.
structure(list(ID = 1:17, a = c(1L, 1L, 1L, 1L, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA), b = c(NA, NA, NA, NA, 2L,
2L, 2L, 2L, 2L, NA, NA, NA, NA, NA, NA, NA, NA), c = c(NA, NA,
NA, NA, NA, NA, NA, NA, NA, 3L, NA, NA, NA, NA, NA, NA, NA),
d = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 4L, NA, NA,
NA, NA, NA, NA), e = c(NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 5L, NA, NA, NA, NA, NA), r = c(NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, 6L, NA, NA, NA, NA), g = c(NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 7L, 7L, 7L,
7L)), row.names = c(NA, -17L), class = c("tbl_df", "tbl",
"data.frame"))
CodePudding user response:
I think it the easiest approach is to convert into a list and remove the blanks, and then put the list back together as a data frame.
dat <- structure(list(ID = 1:17, a = c(1L, 1L, 1L, 1L, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA), b = c(NA, NA, NA, NA, 2L,
2L, 2L, 2L, 2L, NA, NA, NA, NA, NA, NA, NA, NA), c = c(NA, NA,
NA, NA, NA, NA, NA, NA, NA, 3L, NA, NA, NA, NA, NA, NA, NA),
d = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 4L, NA, NA,
NA, NA, NA, NA), e = c(NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 5L, NA, NA, NA, NA, NA), r = c(NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, 6L, NA, NA, NA, NA), g = c(NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 7L, 7L, 7L,
7L)), row.names = c(NA, -17L), class = c("tbl_df", "tbl",
"data.frame"))
dat2 <- list()
for(i in 1:ncol(dat)) { # Using for-loop to add columns to list
x <- dat[ , i]
x <- x[!is.na(x)] #remove the blanks
x <- c(x, rep("", nrow(dat)-length(x))) #make new blanks after the values of interest, as to have equal rows to original
dat2[[i]] <-x
}
res<-data.frame(do.call("cbind", dat2)) #from list to dataframe
names(res) <- names(dat) #put in original names
with the other data structure (very similar)
dat <- data.frame(ID = c(1:17),
"1" = c(rep("a", 4), rep("", 13)),
"2" = c(rep("", 4), rep("b", 5), rep("", 8)),
"3" = c(rep("", 9), "c", rep("", 7)),
"4" = c(rep("", 10), "d", rep("", 6)),
"5" = c(rep("", 11), "e", rep("", 5)),
"6" = c(rep("", 12), "f", rep("", 4)),
"7" = c(rep("", 13), rep("g", 4)))
dat2 <- list()
for(i in 1:ncol(dat)) { # Using for-loop to add columns to list
x <- dat[ , i]
x <- x[x != ""] #remove the blanks
x <- c(x, rep("", nrow(dat)-length(x))) #make new blanks after the values of interest, as to have equal rows to original
dat2[[i]] <-x
}
res<-data.frame(do.call("cbind", dat2)) #from list to dataframe
names(res) <- names(dat) #put in original names
CodePudding user response:
You can use na.last = TRUE. See ?sort
apply(d, 2, sort, na.last = TRUE)
##> ID a b c d e r g
##> [1,] 1 1 2 3 4 5 6 7
##> [2,] 2 1 2 NA NA NA NA 7
##> [3,] 3 1 2 NA NA NA NA 7
##> [4,] 4 1 2 NA NA NA NA 7
##> [5,] 5 NA 2 NA NA NA NA NA
##> [6,] 6 NA NA NA NA NA NA NA
##> [7,] 7 NA NA NA NA NA NA NA
##> [8,] 8 NA NA NA NA NA NA NA
##> [9,] 9 NA NA NA NA NA NA NA
##> [10,] 10 NA NA NA NA NA NA NA
##> [11,] 11 NA NA NA NA NA NA NA
##> [12,] 12 NA NA NA NA NA NA NA
##> [13,] 13 NA NA NA NA NA NA NA
##> [14,] 14 NA NA NA NA NA NA NA
##> [15,] 15 NA NA NA NA NA NA NA
##> [16,] 16 NA NA NA NA NA NA NA
##> [17,] 17 NA NA NA NA NA NA NA
CodePudding user response:
another solution is to filter out each NA, then resize the vectors:
raw <- structure(list(ID = 1:17, a = c(1L, 1L, 1L, 1L, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA), b = c(NA, NA, NA, NA, 2L,
2L, 2L, 2L, 2L, NA, NA, NA, NA, NA, NA, NA, NA), c = c(NA, NA,
NA, NA, NA, NA, NA, NA, NA, 3L, NA, NA, NA, NA, NA, NA, NA),
d = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 4L, NA, NA,
NA, NA, NA, NA), e = c(NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 5L, NA, NA, NA, NA, NA), r = c(NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, 6L, NA, NA, NA, NA), g = c(NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 7L, 7L, 7L,
7L)), row.names = c(NA, -17L), class = c("tbl_df", "tbl",
"data.frame"))
library(tidyverse)
lapply(1:length(raw), function(i){
tmp <- raw[i] %>% filter(!is.na(.)) %>% pull() #filter NA
length(tmp) <- nrow(raw) #resize to length of raw struct
tmp #return
}) -> res
tbl <- res %>% bind_cols() #make table
colnames(tbl) <- raw %>% colnames() #set old column names
tbl
Stefanos solution should be preferred though