I am currently trying to work on a ".dat file" to change some values in the file. I am using
read.delim() to import it into R and it works. But when I want to save the file again, the format changes and I cannot use it anymore. I used write.csv(Tin, file ="TinNew.dat").
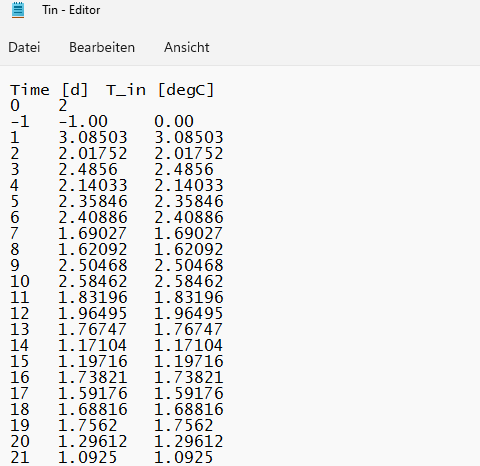
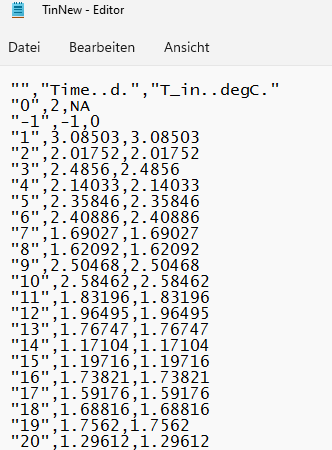
I hope the screenshots help to understand my problem, also I left a snippet of the data, as a reproducible example. I want to have the same format as shown in the "Editor (original)". How can I achieve that?
Thanks in advance
Editor original:
Editor(after I try to save it):
Time [d] T_in [degC]
0 2
-1 -1.00 0.00
1 3.08503 3.08503
2 2.01752 2.01752
3 2.4856 2.4856
4 2.14033 2.14033
5 2.35846 2.35846CodePudding user response:
There are two issues, the header and the separator. Header: It is not practical to have measurement units and column names together in the first row, because it makes creation of the header difficult. It is of course possible in R, but to avoid complication, let's just skip the first row and add the header afterwards.
txt <-
"Time [d] T_in [degC]
0 2
-1 -1.00 0.00
1 3.08503 3.08503
2 2.01752 2.01752
3 2.4856 2.4856
4 2.14033 2.14033
5 2.35846 2.35846"
dat <- read.table(text=txt, skip=1)
names(dat) <- c("Time [d]", "T_in [degC]")
Note that I embedded the data directly in the code and read it from the txt string instead of a file. It works the same with files using read.table(file=filename, skip=1). The default separator of read.table is whitespace, i.e. either space or tab.
To write the file back, use write.table with the following options:
write.table(dat, file="output.dat", sep=" ", quote=FALSE, row.names = FALSE)
Here we use again the space " " as the separator, but it may be better to use a tab "\t" in this case.
More details are found at the help pages, where one can see that read.delim and write.csv are just a convenience functions, i.e. versions of read.tableand write.table with different defaults.
Alternative approach
Finally, we see that row 2 has only two values, so the idea is to handle the two-lines header and the data separately. readLines reads n=2 lines separately as is, and then read.table reads the data. Writing is then done analogously:
header <- readLines("input.dat", n=2)
dat <- read.table(text=txt, skip=2)
writeLines(header, "output.dat")
write.table(dat, "output.dat", sep="\t",
quote=FALSE, col.names=FALSE, row.names = FALSE, append=TRUE)
