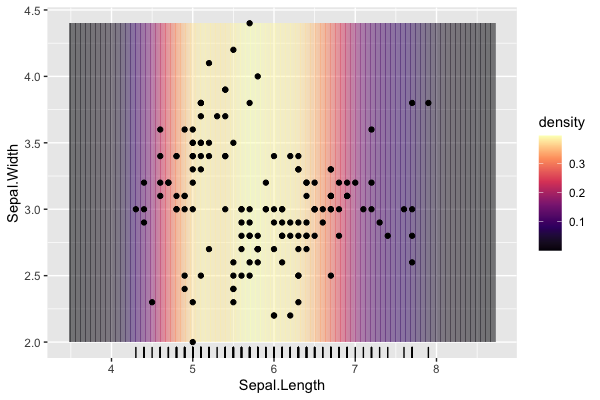
I would like to plot a background that captures the density of points in one dimension in a scatter plot. This would serve a similar purpose to a marginal density plot or a rug plot. I have a way of doing it that is not particularly elegant, I am wondering if there's some built-in functionality I can use to produce this kind of plot.
Mainly there are a few issues with the current approach:
- Alpha overlap at boundaries causes banding at lower resolution as seen here. - Primary objective, looking for a geom or other solution that draws a nice continuous band filled with specific colour. Something like geom_density_2d() but with the stat drawn from only the X axis.
- "Background" does not cover expanded area, can use coord_cartesian(expand = FALSE) but would like to cover regular margins. - Not a big deal, is nice-to-have but not required.
- Setting
scale_fill"consumes" the option for the plot, not allowing it to be set independently for the points themselves. - This may not be easily achievable, independent palettes for layers appears to be a fundamental issue with ggplot2.
data(iris)
dns <- density(iris$Sepal.Length)
dns_df <- tibble(
x = dns$x,
density = dns$y
)%>%
mutate(
start = x - mean(diff(x))/2,
end = x mean(diff(x))/2
)
ggplot()
geom_rect(
data = dns_df,
aes(xmin = start, xmax = end, fill = density),
ymin = min(iris$Sepal.Width),
ymax = max(iris$Sepal.Width),
alpha = 0.5)
scale_fill_viridis_c(option = "A")
geom_point(data = iris, aes(x = Sepal.Length, y = Sepal.Width))
geom_rug(data = iris, aes(x = Sepal.Length))
CodePudding user response:
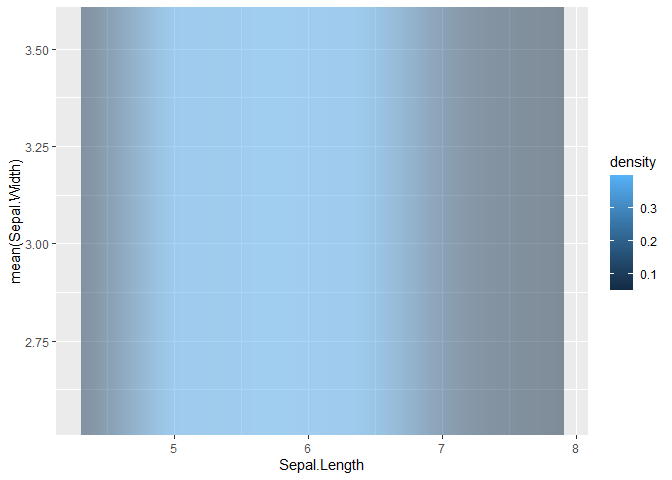
This is a bit of a hacky solution because it (ab)uses knowledge of how objects are internally parametrised to get what you want, which will yield some warnings, but gets you want you'd want.
First, we'll use a geom_raster() stat_density() decorated with some choice after_stat()/stage() delayed evaluation. Normally, this would result in a height = 1 strip, but by setting the internal parameters ymin/ymax to infinitives, we'll have the strip extend the whole height of the plot. Using geom_raster() resolves the alpha issue you were having.
library(ggplot2)
p <- ggplot(iris)
geom_raster(
aes(Sepal.Length,
y = mean(Sepal.Width),
fill = after_stat(density),
ymin = stage(NULL, after_scale = -Inf),
ymax = stage(NULL, after_scale = Inf)),
stat = "density", alpha = 0.5
)
#> Warning: Ignoring unknown aesthetics: ymin, ymax
p
#> Warning: Duplicated aesthetics after name standardisation: NA
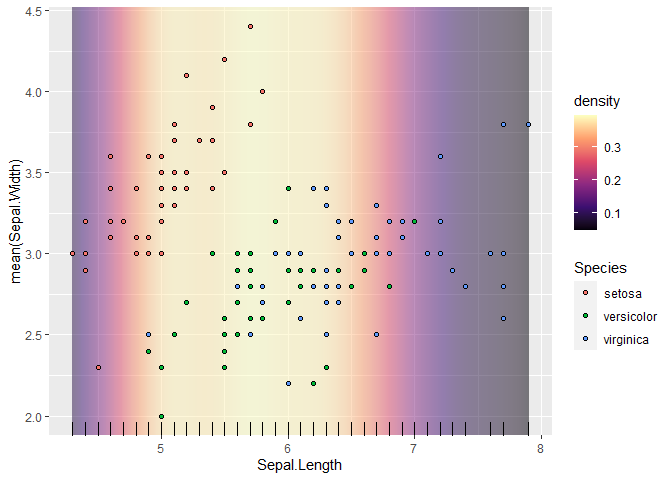
Next, we add a fill scale, and immediately follow that by ggnewscale::new_scale_fill(). This allows another layer to use a second fill scale, as demonstrated with fill = Species.
p <- p
scale_fill_viridis_c(option = "A")
ggnewscale::new_scale_fill()
geom_point(aes(Sepal.Length, Sepal.Width, fill = Species),
shape = 21)
geom_rug(aes(Sepal.Length))
p
#> Warning: Duplicated aesthetics after name standardisation: NA
Lastly, to get rid of the padding at the x-axis, we can manually extend the limits and then shrink in the expansion. It allows for an extended range over which the density can be estimated, making the raster fill the whole area. There is some mismatch between how ggplot2 and scales::expand_range() are parameterised, so the exact values are a bit of trial and error.
p
scale_x_continuous(
limits = ~ scales::expand_range(.x, mul = 0.05),
expand = c(0, -0.2)
)
#> Warning: Duplicated aesthetics after name standardisation: NA
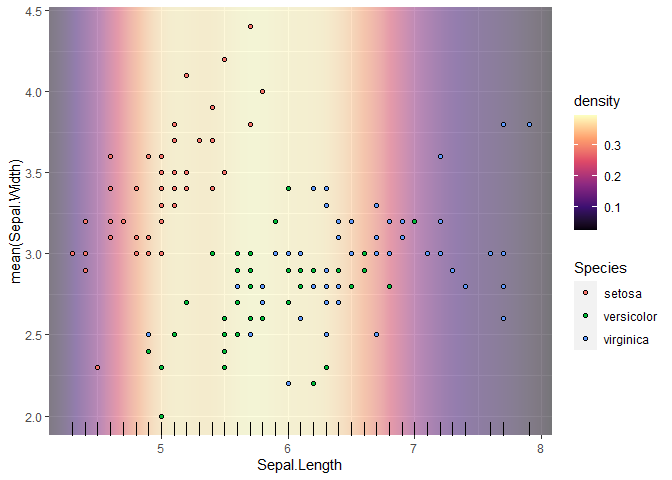
Created on 2022-07-04 by the reprex package (v2.0.1)
CodePudding user response:
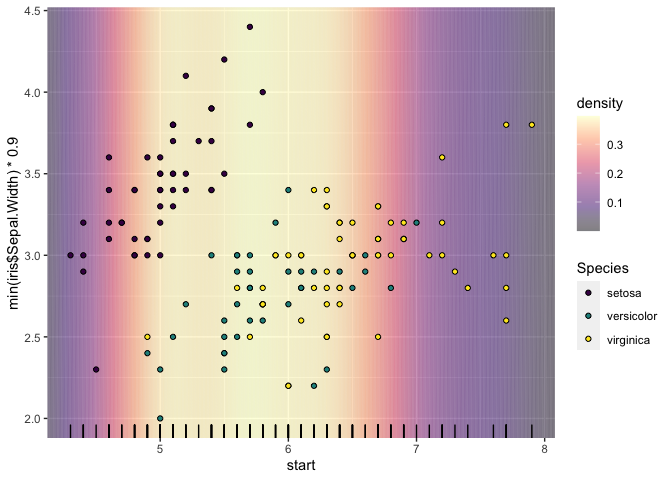
This doesn't solve your problem (I'm not sure I understand all the issues correctly), but perhaps it will help:
- Background does not cover expanded area, can use coord_cartesian(expand = FALSE) but would like to cover regular margins.
If you make the 'background' larger and use coord_cartesian() you can get the same 'filled-to-the-edges' effect; would this work for your use-case?
- Alpha overlap at boundaries causes banding at lower resolution as seen here.
I wasn't able to fix the banding completely, but my approach below appears to reduce it.
- Setting scale_fill "consumes" the option for the plot, not allowing it to be set independently for the points themselves.
If you use geom_segment() you can map density to colour, leaving fill available for e.g. the points. Again, not sure if this is a useable solution, just an idea that might help.
library(tidyverse)
data(iris)
dns <- density(iris$Sepal.Length)
dns_df <- tibble(
x = dns$x,
density = dns$y
) %>%
mutate(
start = x - mean(diff(x))/2,
end = x mean(diff(x))/2
)
ggplot()
geom_segment(
data = dns_df,
aes(x = start, xend = end,
y = min(iris$Sepal.Width) * 0.9,
yend = max(iris$Sepal.Width) * 1.1,
color = density), alpha = 0.5)
coord_cartesian(ylim = c(min(iris$Sepal.Width),
max(iris$Sepal.Width)),
xlim = c(min(iris$Sepal.Length),
max(iris$Sepal.Length)))
scale_color_viridis_c(option = "A", alpha = 0.5)
scale_fill_viridis_d()
geom_point(data = iris, aes(x = Sepal.Length,
y = Sepal.Width,
fill = Species),
shape = 21)
geom_rug(data = iris, aes(x = Sepal.Length))
Created on 2022-07-04 by the reprex package (v2.0.1)