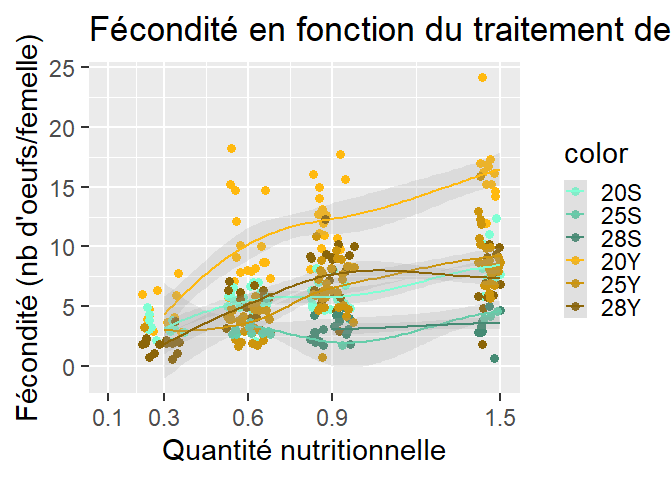
I'm doing a plot with ggplot2 but when I add the function geom_smooth with method = loess, my graph doesn't work . Indeed, it creates curves that don't match with data. When I change this line in my script, by deleting it , or using another method , graphs work correctly.
I already checked that my data are numeric and that there are not extrem values because of a missing decimal, but it is not that.
How can I correct that to make my geom_smooth work with the loess method ?
CodePudding user response:
Your issue is that the default parameters of the loess are not working well for your dataset. You have only a small number of discrete x values so it doesn't know how best to fit it. For example, if you look at the default value of span in base::loess() (which ggplot2::geom_smooth(method = "loess") calls under the hood) you can find that the default value is span = 0.75. If you just increase to span = 0.8 you get what I assume is closer to what you wanted. For more on the span parameter you can see this answer.
library(tidyverse)
d %>%
ggplot(aes(x = quantity, y = fecundity, col = color))
geom_jitter(size = 3)
geom_smooth(method = "loess", span = 0.8, alpha = 0.2)
scale_x_continuous(breaks=c(0.1,0.3,0.6,0.9,1.5), limits=c(0.1,1.5))
scale_colour_manual(values=c("20S" = "aquamarine1","25S" = "aquamarine3","28S" =
"aquamarine4","20Y" = "darkgoldenrod1","25Y" = "darkgoldenrod3", "28Y" = "darkgoldenrod4"))
ggtitle("Fécondité en fonction du traitement de nourriture et de la température")
xlab("Quantité nutritionnelle") ylab("Fécondité (nb d'oeufs/femelle)")
theme_grey(base_size = 22)
Created on 2022-07-05 by the reprex package (v2.0.1)
CodePudding user response:
You need to quote the method type, e.g. use geom_smooth(method="loess")
