I've a text file containg some data for correlation function. It is structured like 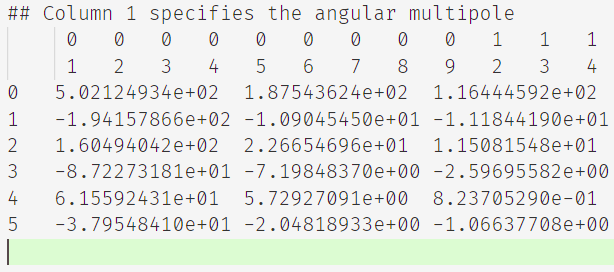
The first two rows are bin numbers and 45 entries, while the remaining rows are the values at the given location and containers 46 entries in each row. In these remaining rows, i.e, from row 2 , the first column is the order of the values.
I want to read this as a pandas data frame. Since there is a mismatch of dimension, pandas show an error.
ParserError: Error tokenizing data. C error: Expected 45 fields in line 9, saw 46
To fix this error, I modify the txt file by adding 'r1' and 'r2' in place of the blank space in the first two rows.
This is a solution if there are only a few files, but unfortunately, I've 100s of files structured in the same way. Is there a way to read the data from this. It would be fine for me if I can just skip first column entirely from row two onwards.
CodePudding user response:
I may would try to read at first all rows except the first two with the skiprows attribute in the read_csv() function. Afterwards read the first two rows with the nrows=2 attribute.
But thats only a workaround. Maybe there are better solutions.
In the end combine your given dataframes.
Example:
import pandas as pd
rows_uneven_dimensions = 2
df_first_two_rows = pd.read_csv("test.txt", header=None, nrows=rows_uneven_dimensions, sep='\t')
df_all_other_rows = pd.read_csv("test.txt", header=None, skiprows=rows_uneven_dimensions, sep='\t')
frames = [df_first_two_rows, df_all_other_rows]
result = pd.concat(frames, ignore_index=True, axis=0)
test.txt
0 0 0 0 0 0 0
1 2 2 3 4 5 6
0 A B C D E F G
1 H I J K L M N
2 O P Q R S T U
3 V W X Y Z A B
result dataframe value:
[[0 0 0 0 0 0 0 nan]
[1 2 2 3 4 5 6 nan]
[0 'A' 'B' 'C' 'D' 'E' 'F' 'G']
[1 'H' 'I' 'J' 'K' 'L' 'M' 'N']
[2 'O' 'P' 'Q' 'R' 'S' 'T' 'U']
[3 'V' 'W' 'X' 'Y' 'Z' 'A' 'B']]
The last value in the first two rows gets filled with nan by default in the pd.concat function.
CodePudding user response:
It seems like the two first rows are some kind of Multiindex-Columns, thus the entries of the row-index are missing (45 instead of 46 columns).
If my guess is correct you can extend Oivalfs answer by pandas Multiindex:
import pandas as pd
rows_uneven_dimensions = 2
df_first_two_rows = pd.read_csv("test.txt", header=None, nrows=rows_uneven_dimensions, sep='\t')
df_all_other_rows = pd.read_csv("test.txt", header=None, skiprows=rows_uneven_dimensions, sep='\t', index_col=0) #Defining first col as index
df_all_other_rows.index.name = 'Index' # Optional: Set index Name
cols=pd.MultiIndex.from_arrays([df_first_two_rows.iloc[idx] for idx in df_first_two_rows.index], names=("First Level", "Second Level")) # Define Multinidex based on Rows of df_first_two_rows
# Names of Levels is just for illustration purpose
df_all_other_rows.columns=cols # Replacing the old col-index with new Multiindex
print(df_all_other_rows)
Given the test.txt given by Oivalf the result will look like this:
First Level 0
Second Level 1 2 2 3 4 5 6
Index
0 A B C D E F G
1 H I J K L M N
2 O P Q R S T U
3 V W X Y Z A B
