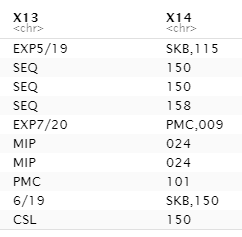
I have raw data that when importing, there is some "extra" data for some of the rows. It is saved as a text file but is comma separated. When bringing it into R, it looks like this:
In the pic above, where you see "EXP5/19", "EXP7/20", "6/19" in X13 is extra data that only some rows have. When using read_csv it pushes the data that's supposed to be in column 13 into column 14. When using read.csv, it makes the value in the last column as the value in the first column of the next observation.
read_csv("filename.txt", col_names = F)
read.csv("filename.txt", header=F, stringsAsFactors=F)
There are close to 400 of these rows, so I can open WordPad and manually remove them. Anyway I can do it programmatically in R?
CodePudding user response:
are you looking for something like this?
df <- data.frame(X13 = c("EXP5/19", "SEQ", "SEQ", "SEQ", "EXP7/20"),
X14 = c("SKB,115", 150, 150, 158, "PMC,009"))
library(dplyr)
library(stringr)
df %>%
# if "," in X14 extract leading characters as X13
dplyr::mutate(X13 = ifelse(stringr::str_detect(X14, pattern = ","),
stringr::str_extract(X14, pattern = "^[:alpha:]*"),
X13),
# remove leading characters incl. comma
X14 = stringr::str_remove(X14, pattern = "^[:alpha:]*,"))
X13 X14
1 SKB 115
2 SEQ 150
3 SEQ 150
4 SEQ 158
5 PMC 009