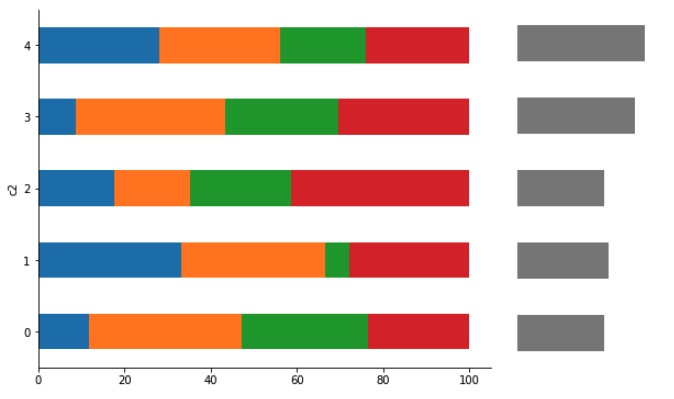
I have the following plot
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
fig = plt.figure(figsize=(10, 6))
ax_dict = fig.subplot_mosaic(
[
["a", "a", "a", "b"],
["a", "a", "a", "b"],
["a", "a", "a", "b"],
["a", "a", "a", "b"],
]
)
rng = np.random.default_rng(1)
df = pd.DataFrame(
{
"c1": rng.integers(0, 4, 100),
"c2": rng.integers(0, 5, 100),
}
)
ax = ax_dict["a"]
pd.crosstab(df["c1"], df["c2"], normalize="columns").mul(100).round(1).T.plot.barh(
stacked=True, ax=ax
)
stacked_bar_patch = ax.patches[0]
ax.get_legend().remove()
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax = ax_dict["b"]
counts = df["c2"].value_counts()
ax.barh(
counts.index,
counts,
color="grey",
height=stacked_bar_patch.get_height(),
)
_ = ax.set_xticks([])
_ = ax.set_yticks([])
ax.spines["left"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.spines["bottom"].set_visible(False)
Which looks as :
I would like to arrange the patches on the right plot (ax_dict['b']) so that they're horizontally aligned with the bars from ax_dict['a'].
Currently they're roughly inline - but the bars are higher / lower on the right than the bars on the left (red circles indicate the "gaps", which wouldn't be present if they were lined up exactly):
My question is - how can i create this plot so that the bars line up exactly ?
CodePudding user response:
This issue can easily be fixed by giving both axes the same y limits, in your example this might be done simply by adding:
ax.set_ylim(-0.5,4.5)
for each subplot, i.e. once before and once after the line ax = ax_dict["b"].
Sidenote: you should consider storing each unique axis within a unique variable to avoid confusion, e.g. naming them ax_a and ax_b.