I would like to create a bar plot using R, from my GO term of several genes. In the GO results, there were 3 categories which is Term, Module and Adjusted.P.value.
Here's my data file:
Term,Module,Adjusted P-value
interferon-gamma-mediated signaling pathway (GO:0060333),Biological Process,1.57E-10
antigen processing and presentation of exogenous peptide antigen (GO:0002478),Biological Process,1.57E-10
cytokine-mediated signaling pathway (GO:0019221),Biological Process,2.89E-10
antigen processing and presentation of exogenous peptide antigen via MHC class II (GO:0019886),Biological Process,2.11E-09
antigen processing and presentation of peptide antigen via MHC class II (GO:0002495),Biological Process,2.11E-09
cellular response to interferon-gamma (GO:0071346),Biological Process,9.98E-09
antigen receptor-mediated signaling pathway (GO:0050851),Biological Process,1.61E-08
T cell receptor signaling pathway (GO:0050852),Biological Process,8.20E-08
rRNA processing (GO:0006364),Biological Process,3.61E-06
maturation of 5.8S rRNA (GO:0000460),Biological Process,1.45E-05
maturation of LSU-rRNA (GO:0000470),Biological Process,8.13E-05
ribosome biogenesis (GO:0042254),Biological Process,1.05E-04
ncRNA processing (GO:0034470),Biological Process,1.31E-04
positive regulation of leukocyte cell-cell adhesion (GO:1903039),Biological Process,1.33E-04
positive regulation of T cell mediated immunity (GO:0002711),Biological Process,2.30E-04
toll-like receptor signaling pathway (GO:0002224),Biological Process,3.96E-04
rRNA metabolic process (GO:0016072),Biological Process,3.96E-04
positive regulation of leukocyte mediated cytotoxicity (GO:0001912),Biological Process,3.96E-04
antigen processing and presentation of endogenous peptide antigen (GO:0002483),Biological Process,4.35E-04
positive regulation of nucleic acid-templated transcription (GO:1903508),Biological Process,6.91E-04
MHC class II receptor activity (GO:0032395),Molecular Functions,6.30E-09
RNA binding (GO:0003723),Molecular Functions,5.00E-04
RNA polymerase II-specific DNA-binding transcription factor binding (GO:0061629),Molecular Functions,8.84E-04
complement component C3b binding (GO:0001851),Molecular Functions,0.00284344
protein kinase binding (GO:0019901),Molecular Functions,0.00284344
DNA-binding transcription factor binding (GO:0140297),Molecular Functions,0.007423848
kinase binding (GO:0019900),Molecular Functions,0.007423848
I-SMAD binding (GO:0070411),Molecular Functions,0.012276789
methylation-dependent protein binding (GO:0140034),Molecular Functions,0.013810415
methylated histone binding (GO:0035064),Molecular Functions,0.015454238
MHC class II protein complex binding (GO:0023026),Molecular Functions,0.015454238
bHLH transcription factor binding (GO:0043425),Molecular Functions,0.023843033
lumenal side of endoplasmic reticulum membrane (GO:0098553),Cellular Component,2.77E-15
integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556),Cellular Component,2.77E-15
MHC protein complex (GO:0042611),Cellular Component,1.40E-14
ER to Golgi transport vesicle membrane (GO:0012507),Cellular Component,9.63E-13
coated vesicle membrane (GO:0030662),Cellular Component,9.63E-13
transport vesicle membrane (GO:0030658),Cellular Component,1.85E-12
MHC class II protein complex (GO:0042613),Cellular Component,1.29E-11
COPII-coated ER to Golgi transport vesicle (GO:0030134),Cellular Component,1.85E-11
endocytic vesicle membrane (GO:0030666),Cellular Component,3.25E-10
integral component of endoplasmic reticulum membrane (GO:0030176),Cellular Component,3.13E-09
clathrin-coated endocytic vesicle membrane (GO:0030669),Cellular Component,1.17E-08
endocytic vesicle (GO:0030139),Cellular Component,3.30E-08
clathrin-coated endocytic vesicle (GO:0045334),Cellular Component,4.37E-08
clathrin-coated vesicle membrane (GO:0030665),Cellular Component,6.07E-08
cytoplasmic vesicle membrane (GO:0030659),Cellular Component,7.43E-08
trans-Golgi network membrane (GO:0032588),Cellular Component,1.04E-07
lytic vacuole membrane (GO:0098852),Cellular Component,6.24E-06
lysosome (GO:0005764),Cellular Component,6.24E-06
bounding membrane of organelle (GO:0098588),Cellular Component,9.26E-06
lysosomal membrane (GO:0005765),Cellular Component,2.70E-05
When using the code below, I was able to generate a bar chart.
ggplot(data, aes(x = Term, y = -log10(Adjusted.P.value), fill = Module)) geom_bar(stat = "identity", position = "dodge") coord_flip()
However, the x-axis of the bar chart was scattered around, instead of grouping together. By "grouping together", reds should be together, greens should be together etc etc..
Could anyone please advise or revise my code?
Thank you so much.
CodePudding user response:
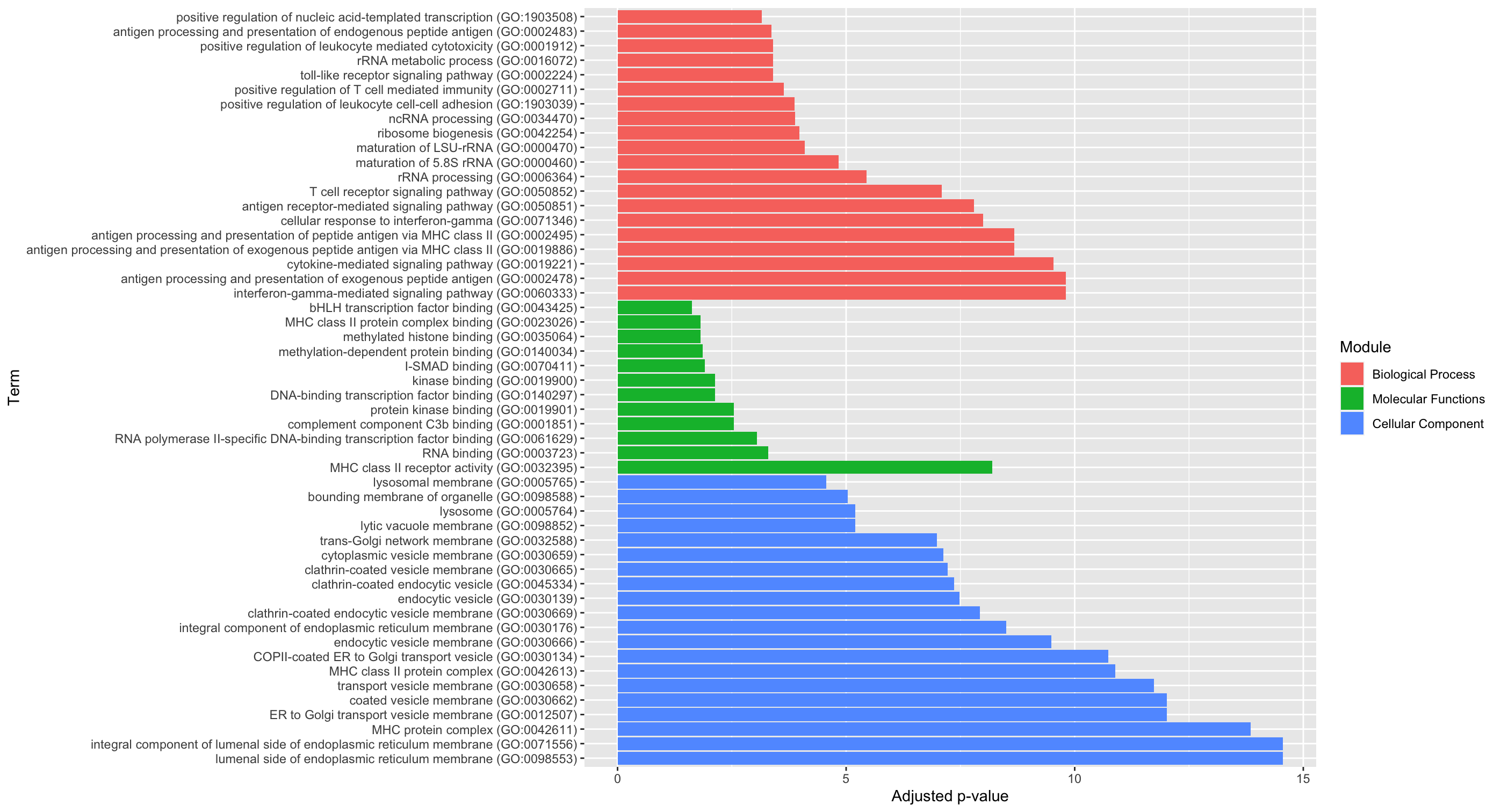
To order by the Module, we can make Module a factor, then create the order with levels and rearrange by the desired order. Then, you can use fct_inorder from forcats (part of the tidyverse) to keep the order that we have arranged by. Here, I also use fct_rev on fill to match the top down order for the legend.
library(tidyverse)
data %>%
mutate(Module = factor(Module, levels = c("Cellular Component", "Molecular Functions", "Biological Process"))) %>%
arrange(Module) %>%
ggplot(aes(x = fct_inorder(Term), y = -log10(Adjusted.P.value), fill = fct_rev(Module)))
geom_bar(stat = "identity", position = "dodge")
coord_flip()
xlab("Term")
ylab("Adjusted p-value")
guides(fill=guide_legend(title = "Module"))
Output
Or another option (if it is already in order) would be to plot by the row number for your x. Then, we can use labels in scale_x_discrete to put the Term instead of the row number along the axis.
ggplot(data, aes(x = 1:nrow(data), y = -log10(Adjusted.P.value), fill = Module))
geom_bar(stat = "identity", position = "dodge")
scale_x_discrete(labels=data$Term, breaks=1:nrow(data), limits=factor(1:nrow(data)), name='Term')
coord_flip()
Data
data <- structure(list(Term = c("interferon-gamma-mediated signaling pathway (GO:0060333)",
"antigen processing and presentation of exogenous peptide antigen (GO:0002478)",
"cytokine-mediated signaling pathway (GO:0019221)", "antigen processing and presentation of exogenous peptide antigen via MHC class II (GO:0019886)",
"antigen processing and presentation of peptide antigen via MHC class II (GO:0002495)",
"cellular response to interferon-gamma (GO:0071346)", "antigen receptor-mediated signaling pathway (GO:0050851)",
"T cell receptor signaling pathway (GO:0050852)", "rRNA processing (GO:0006364)",
"maturation of 5.8S rRNA (GO:0000460)", "maturation of LSU-rRNA (GO:0000470)",
"ribosome biogenesis (GO:0042254)", "ncRNA processing (GO:0034470)",
"positive regulation of leukocyte cell-cell adhesion (GO:1903039)",
"positive regulation of T cell mediated immunity (GO:0002711)",
"toll-like receptor signaling pathway (GO:0002224)", "rRNA metabolic process (GO:0016072)",
"positive regulation of leukocyte mediated cytotoxicity (GO:0001912)",
"antigen processing and presentation of endogenous peptide antigen (GO:0002483)",
"positive regulation of nucleic acid-templated transcription (GO:1903508)",
"MHC class II receptor activity (GO:0032395)", "RNA binding (GO:0003723)",
"RNA polymerase II-specific DNA-binding transcription factor binding (GO:0061629)",
"complement component C3b binding (GO:0001851)", "protein kinase binding (GO:0019901)",
"DNA-binding transcription factor binding (GO:0140297)", "kinase binding (GO:0019900)",
"I-SMAD binding (GO:0070411)", "methylation-dependent protein binding (GO:0140034)",
"methylated histone binding (GO:0035064)", "MHC class II protein complex binding (GO:0023026)",
"bHLH transcription factor binding (GO:0043425)", "lumenal side of endoplasmic reticulum membrane (GO:0098553)",
"integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556)",
"MHC protein complex (GO:0042611)", "ER to Golgi transport vesicle membrane (GO:0012507)",
"coated vesicle membrane (GO:0030662)", "transport vesicle membrane (GO:0030658)",
"MHC class II protein complex (GO:0042613)", "COPII-coated ER to Golgi transport vesicle (GO:0030134)",
"endocytic vesicle membrane (GO:0030666)", "integral component of endoplasmic reticulum membrane (GO:0030176)",
"clathrin-coated endocytic vesicle membrane (GO:0030669)", "endocytic vesicle (GO:0030139)",
"clathrin-coated endocytic vesicle (GO:0045334)", "clathrin-coated vesicle membrane (GO:0030665)",
"cytoplasmic vesicle membrane (GO:0030659)", "trans-Golgi network membrane (GO:0032588)",
"lytic vacuole membrane (GO:0098852)", "lysosome (GO:0005764)",
"bounding membrane of organelle (GO:0098588)", "lysosomal membrane (GO:0005765)"
), Module = c("Biological Process", "Biological Process", "Biological Process",
"Biological Process", "Biological Process", "Biological Process",
"Biological Process", "Biological Process", "Biological Process",
"Biological Process", "Biological Process", "Biological Process",
"Biological Process", "Biological Process", "Biological Process",
"Biological Process", "Biological Process", "Biological Process",
"Biological Process", "Biological Process", "Molecular Functions",
"Molecular Functions", "Molecular Functions", "Molecular Functions",
"Molecular Functions", "Molecular Functions", "Molecular Functions",
"Molecular Functions", "Molecular Functions", "Molecular Functions",
"Molecular Functions", "Molecular Functions", "Cellular Component",
"Cellular Component", "Cellular Component", "Cellular Component",
"Cellular Component", "Cellular Component", "Cellular Component",
"Cellular Component", "Cellular Component", "Cellular Component",
"Cellular Component", "Cellular Component", "Cellular Component",
"Cellular Component", "Cellular Component", "Cellular Component",
"Cellular Component", "Cellular Component", "Cellular Component",
"Cellular Component"), Adjusted.P.value = c(1.57e-10, 1.57e-10,
2.89e-10, 2.11e-09, 2.11e-09, 9.98e-09, 1.61e-08, 8.2e-08, 3.61e-06,
1.45e-05, 8.13e-05, 0.000105, 0.000131, 0.000133, 0.00023, 0.000396,
0.000396, 0.000396, 0.000435, 0.000691, 6.3e-09, 5e-04, 0.000884,
0.00284344, 0.00284344, 0.007423848, 0.007423848, 0.012276789,
0.013810415, 0.015454238, 0.015454238, 0.023843033, 2.77e-15,
2.77e-15, 1.4e-14, 9.63e-13, 9.63e-13, 1.85e-12, 1.29e-11, 1.85e-11,
3.25e-10, 3.13e-09, 1.17e-08, 3.3e-08, 4.37e-08, 6.07e-08, 7.43e-08,
1.04e-07, 6.24e-06, 6.24e-06, 9.26e-06, 2.7e-05)), class = "data.frame", row.names = c(NA,
-52L))