I've recently written my first ggplot2 stat and geom methods. I want to write another that uses the data passed in ggplot2::ggplot(data=) to add a p-value as a caption to the figure. Is that possible?
For example, I would like to write something like this:
library(ggplot2)
mtcars |>
ggplot(aes(x = mpg, y = cyl))
add_pvalue()
Where add_pvalue() would calculate a p-value (e.g. an anova p-value for different mean MPG by the number of cylinders), and add the p-value as a caption, labs(caption = "p = 0.45").
Thank you!
CodePudding user response:
labs(caption = paste("P Value =", pvalue))
Is this what you want?
CodePudding user response:
Daniel, it's possible. You can use this example. Hope that help you !
library(ggplot2)
library(glue)
p_value <- 0.05
mtcars |>
ggplot(aes(x = mpg, y = cyl))
labs(caption = glue("p = {p_value}"))
CodePudding user response:
You could do something like the following, picking your preferred statistical model, "types" of p-values, and formatting of the p-value. If you wanted to build in lots of functionality to make it useful for a wide variety of models, you'd want to add conditional extractor functions for those models.
# Packages
library(ggplot2)
library(dplyr)
library(rlang)
# Define "add_pvalue()" function
# adds p-value from linear regression of y on x
# note that this assumes x and y are reals or integers
add_pvalue <- function(ggplot_obj) {
# Get x and y variable names from ggplot object
x <- ggplot_obj$mapping$x %>%
rlang::quo_get_expr() %>%
deparse()
y <- ggplot_obj$mapping$y %>%
rlang::quo_get_expr() %>%
deparse()
# Build regression model formula, fit model, return model summary
mod <- paste0(y, "~ ", x) %>%
as.formula() %>%
lm(data = ggplot_obj$data) %>%
summary()
# Extract two-tailed t-test p-value from model object (reformat as desired)
pval <- mod$coefficients[x, "Pr(>|t|)"]
# Add p_value as plot caption
ggplot_obj
labs(caption = paste0("p = ", pval))
}
# Example with p-value for linear model and 95% confidence intervals
mtcars %>%
ggplot(aes(x = mpg, y = cyl)) %>%
add_pvalue()
geom_smooth(method = "lm", se = TRUE, level = 0.95)
#> `geom_smooth()` using formula 'y ~ x'
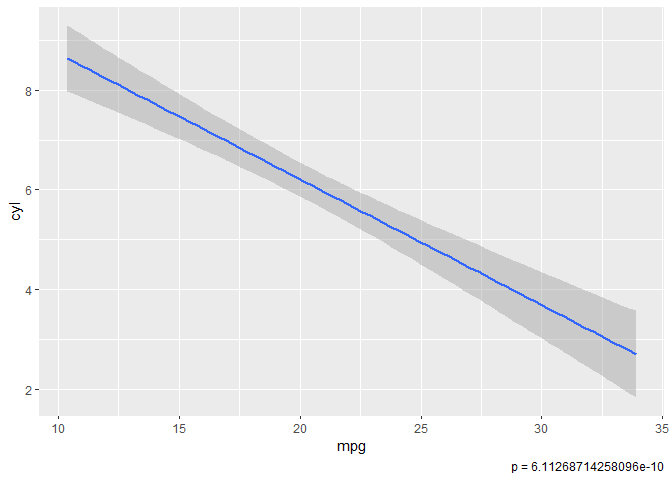
Note that blindly fitting a linear regression or ANOVA to your data is probably not the best decision since x or y may not be real or integer types. If they aren't, this won't really make sense since some models either throw runtime errors or employ one-hot encoding when passed other types of variables.
Similarly, the p-values you obtain may be utterly meaningless if, for example, each row in the data is not an independent observation, you run lots of models that invalidate the sampling assumptions of p-values, your hypothesis doesn't match the test, etc.
Finally, you could also try using the output of stat_smooth() that is produced when you call geom_smooth() to do this. The upside would be that you wouldn't need to fit the model twice to have both that geom and the p-value (using the standard error and coefficients plus normal distribution to get the p-value). That's a bit outside of the scope and would be more limiting since you're stuck with the models it employs and the same issues plague those as well. It's also pretty annoying to extract those: Method to extract stat_smooth line fit
