I am playing with tidymodels workflow for ctree with new bonsai package, an extension for modeling with partykit, here is my code:
pacman::p_load(tidymodels, bonsai, modeldata, finetune)
data(penguins)
doParallel::registerDoParallel()
split <- initial_split(penguins, strata = species)
df_train <- training(split)
df_test <- testing(split)
folds <-
# vfold_cv(train, strata = penguins)
bootstraps(df_train, strata = species, times = 5) # if small number of records
tree_recipe <-
recipe(formula = species ~ flipper_length_mm island, data = df_train)
tree_spec <-
decision_tree(
tree_depth = tune(),
min_n = tune()
) %>%
set_engine("partykit") %>%
set_mode("classification")
tree_workflow <-
workflow() %>%
add_recipe(tree_recipe) %>%
add_model(tree_spec)
set.seed(8833)
tree_tune <-
tune_sim_anneal(
tree_workflow,
resamples = folds,
iter = 30,
initial = 4,
metrics = metric_set(roc_auc, pr_auc, accuracy))
final_workflow <- finalize_workflow(tree_workflow, select_best(tree_tune, "roc_auc"))
final_fit <- last_fit(final_workflow, split = split)
I understand that to extract a final fit model I need to:
final_model <- extract_fit_parsnip(final_fit)
And then I can plot the tree.
plot(final_model$fit)
I would like to try a different plotting library that works with partykit:
library(ggparty)
ggparty(final_model$fit)
geom_edge()
geom_edge_label()
geom_node_splitvar()
geom_node_plot(
gglist = list(geom_bar(x = "", color = species),
xlab("species")),
# draw individual legend for each plot
shared_legend = FALSE
)
But the ggparty code works up to the last line (without it the tree looks good, it prints without plots in final nodes).
It does not see the data inside the fitted model, namely, the response variable species.
Error in layer(data = data, mapping = mapping, stat = stat, geom = GeomBar, :
object 'species' not found
How can I extract the final fit from tidymodels, so that it contains the fitted values as it would if I had built a model without tidymodels workflow?
CodePudding user response:
There are two problems in your code, only one of them related to tidymodels.
The arguments to
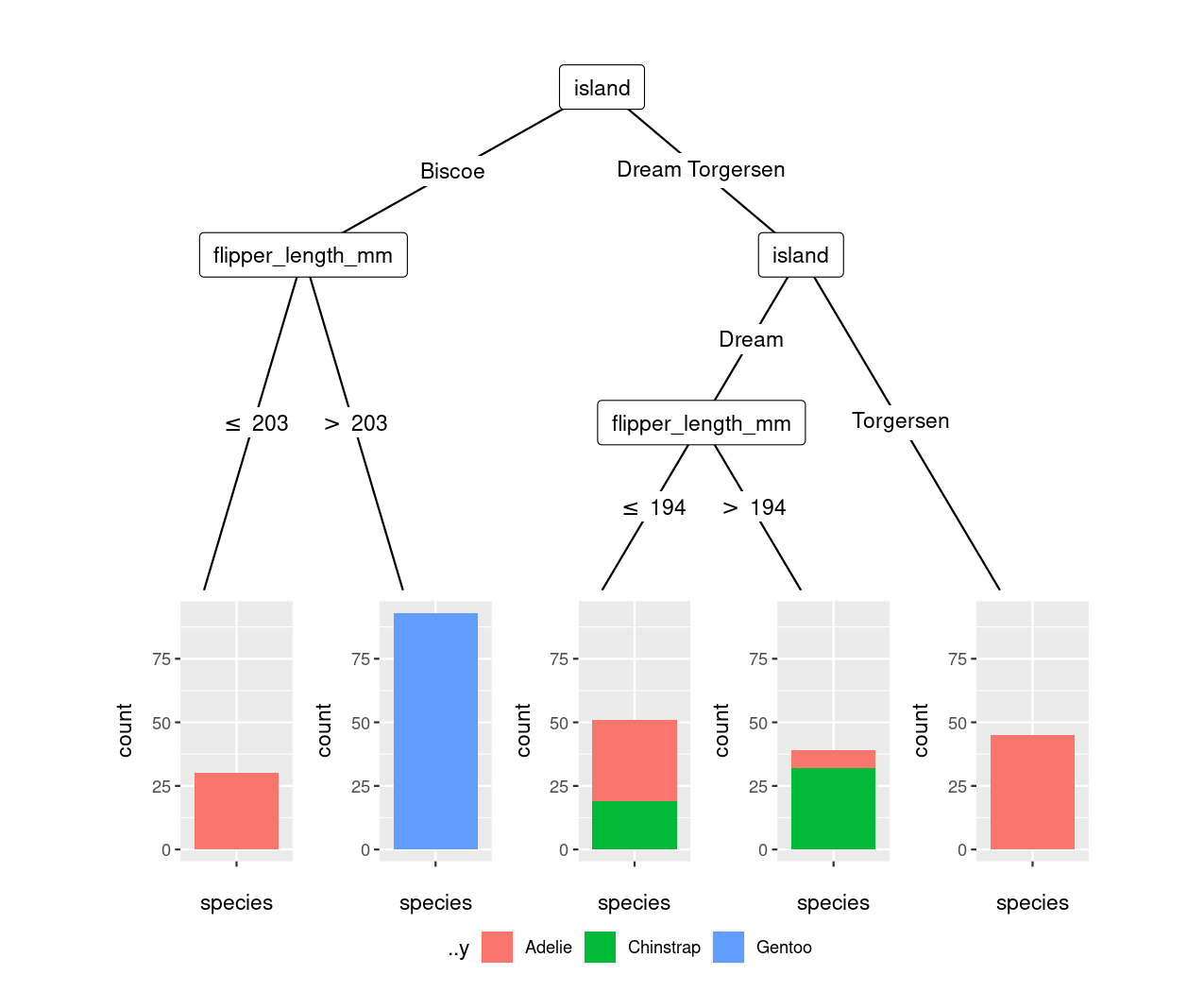
geom_bar()need to be wrapped inaes(), which is necessary both for plainctree()output and for the result from thetidymodelsworkflow.The dependent variable in the output from the
tidymodelsworkflow is not calledspeciesanymore but..y(presumably a standardized placeholder employed intidymodels). This can be seen from simply printing the object:final_model$fit ## Model formula: ## ..y ~ flipper_length_mm island ## ## Fitted party: ## [1] root ## ...
Addressing both of these (plus using the fill= instead of color= aesthetic) works as intended. (Bonus comment: autoplot(final_model$fit) also just works!)
ggparty(final_model$fit)
geom_edge()
geom_edge_label()
geom_node_splitvar()
geom_node_plot(gglist = list(
geom_bar(aes(x = "", fill = ..y)),
xlab("species")
))