I have a phyloseq object containg an otu_table, tax_table and sam_data.physeq is the phyloseq object.
I want to use following code:
plot_richness(physeq, x ="Location" , measures = c("Observed", "Shannon")) geom_boxplot()
Location is a column in sam_data with location 1,2,3 and 4, with each of my 100 samples associated with one location.
This is the plot i'm getting: One boxplot for observed and one for shannon. I want 4 boxplots in both.
I also keep geting a warning:
Warning message:
Continuous x aesthetic -- did you forget aes(group=...)?
I have seen identical code with the correct result without using aes.
CodePudding user response:
The warning message tells you that Location is a continuous numeric variable. You should convert Location to a factor or character before calling plot_richness Here's an example using GlobalPatterns.
require("phyloseq")
require("ggplot2")
# Load data
data("GlobalPatterns")
sample_data(GlobalPatterns)$Location <- sample(c(1, 2, 3, 4), nsamples(GlobalPatterns), replace = T)
plot_richness(GlobalPatterns, x = "Location", measures = c("Shannon", "Observed"))
geom_boxplot()
#> Warning: Continuous x aesthetic -- did you forget aes(group=...)?
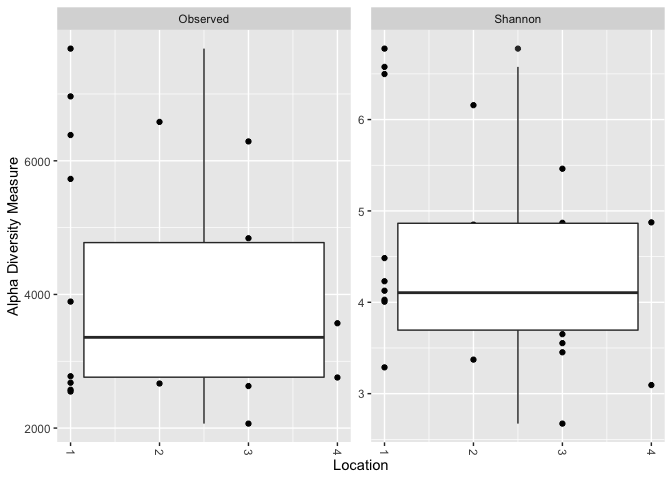
# Update to factor
sample_data(GlobalPatterns)$Location <- as.factor(sample_data(GlobalPatterns)$Location)
plot_richness(GlobalPatterns, x = "Location", measures = c("Shannon", "Observed"))
geom_boxplot()
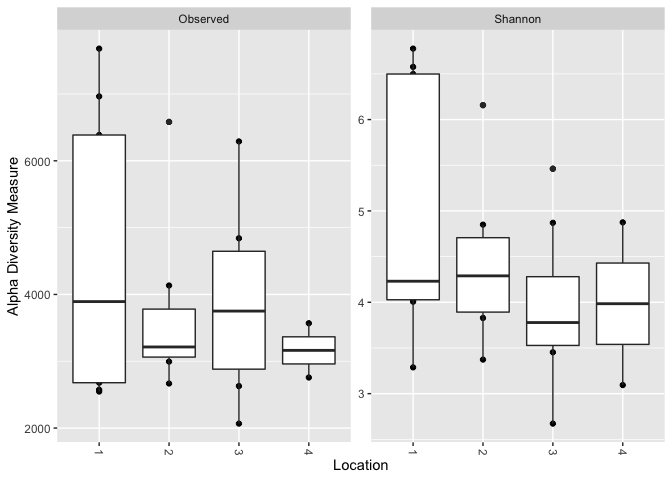
Created on 2022-09-08 by the reprex package (v2.0.1)
