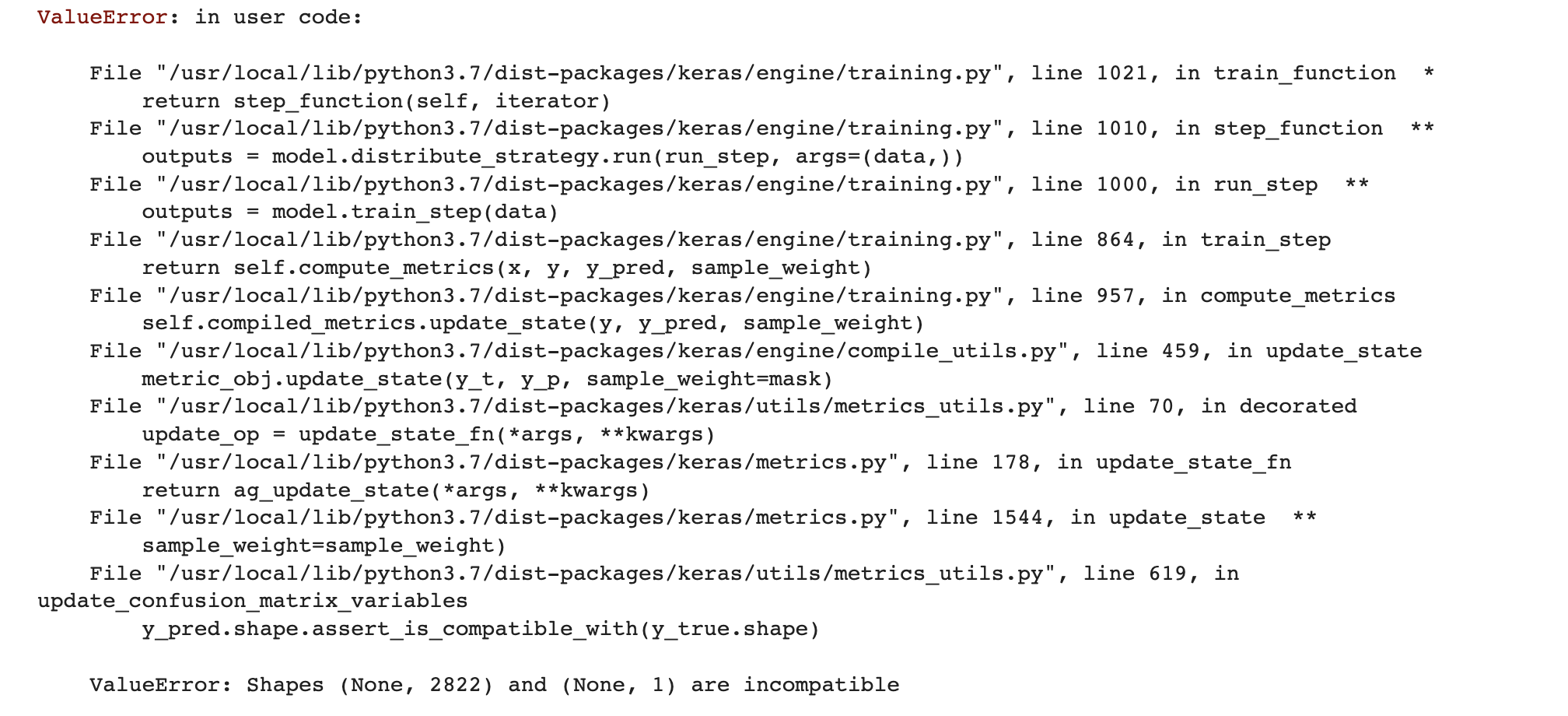
I am building a Siamese network using Keras(TensorFlow) where the target is a binary column, i.e., match or mismatch(1 or 0). But the model fit method throws an error saying that the y_pred is not compatible with the y_true shape. I am using the binary_crossentropy loss function.
Here is the error I see:
Here is the code I am using:
model.compile(loss='binary_crossentropy', optimizer=optimizer, metrics=[tf.keras.metrics.Recall()])
history = model.fit([X_train_entity_1.todense(),X_train_entity_2.todense()],np.array(y_train),
epochs=2,
batch_size=32,
verbose=2,
shuffle=True)
My Input data shapes are as follows:
Inputs:
X_train_entity_1.shape is (700,2822)
X_train_entity_2.shape is (700,2822)
Target:
y_train.shape is (700,1)
In the error it throws, y_pred is the variable which was created internally. What is y_pred dimension is 2822 when I am having a binary target. And 2822 dimension actually matches the input size, but how do I understand this?
Here is the model I created:
in_layers = []
out_layers = []
for i in range(2):
input_layer = Input(shape=(1,))
embedding_layer = Embedding(embed_input_size 1, embed_output_size)(input_layer)
lstm_layer_1 = Bidirectional(LSTM(1024, return_sequences=True,recurrent_dropout=0.2, dropout=0.2))(embedding_layer)
lstm_layer_2 = Bidirectional(LSTM(512, return_sequences=True,recurrent_dropout=0.2, dropout=0.2))(lstm_layer_1)
in_layers.append(input_layer)
out_layers.append(lstm_layer_2)
merge = concatenate(out_layers)
dense1 = Dense(256, activation='relu', kernel_initializer='he_normal', name='data_embed')(merge)
drp1 = Dropout(0.4)(dense1)
btch_norm1 = BatchNormalization()(drp1)
dense2 = Dense(32, activation='relu', kernel_initializer='he_normal')(btch_norm1)
drp2 = Dropout(0.4)(dense2)
btch_norm2 = BatchNormalization()(drp2)
output = Dense(1, activation='sigmoid')(btch_norm2)
model = Model(inputs=in_layers, outputs=output)
model.summary()
Since my data is very sparse, I used todense. And there the type is as follows:
type(X_train_entity_1) is scipy.sparse.csr.csr_matrix
type(X_train_entity_1.todense()) is numpy.matrix
type(X_train_entity_2) is scipy.sparse.csr.csr_matrix
type(X_train_entity_2.todense()) is numpy.matrix
Summary of last few layers as follows:

CodePudding user response:
Mismatched shape in the Input layer. The input shape needs to match the shape of a single element passed as x, or dataset.shape[1:]. So since your dataset size is (700,2822), that is 700 samples of size 2822. So your input shape should be 2822.
Change:
input_layer = Input(shape=(1,))
To:
input_layer = Input(shape=(2822,))
CodePudding user response:
You need to set return_sequences in the lstm_layer_2 to False:
lstm_layer_2 = Bidirectional(LSTM(512, return_sequences=False, recurrent_dropout=0.2, dropout=0.2))(lstm_layer_1)
Otherwise, you will still have the timesteps of your input. That is why you have the shape (None, 2822, 1). You can also add a Flatten layer prior to your output layer, but I would recommend setting return_sequences=False.
Note that a Dense layer computes the dot product between the inputs and the kernel along the last axis of the inputs.