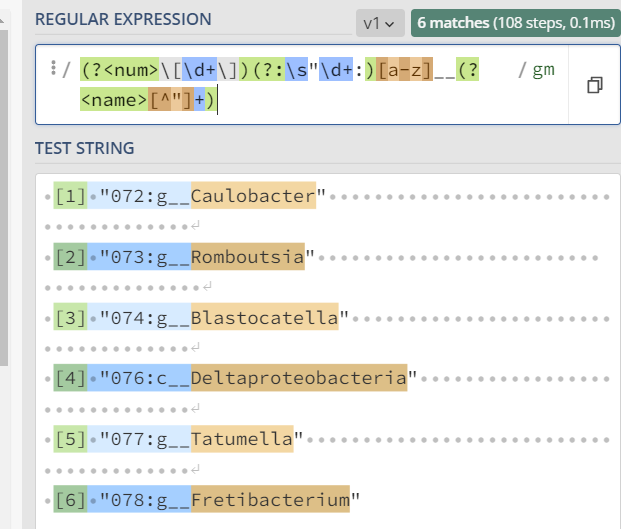
I have a character vector that looks like this:
my_vec
[1] "072:g__Caulobacter"
[2] "073:g__Romboutsia"
[3] "074:g__Blastocatella"
[4] "076:c__Deltaproteobacteria"
[5] "077:g__Tatumella"
[6] "078:g__Fretibacterium"
I want to cut the prefix so the result is the following:
[1] "Caulobacter"
[2] "Romboutsia"
[3] "Blastocatella"
[4] "Deltaproteobacteria"
[5] "tatumella"
[6] "Fretibacterium"
I think using regexps is the way to do this but I'm not familiar with how to do this. The common pattern is the double __.
CodePudding user response:
You can use word from stringr:
library(stringr)
word(my_vec, 2, sep = "__")
#[1] "Caulobacter" "Romboutsia" "Blastocatella"
#[4] "Deltaproteobacteria" "Tatumella" "Fretibacterium"
Another option is to use substring, where regexpr provides the position for __, then we use substring to get the rest of the word by using the starting position of 2 (the first letter after the underscores) to the end of the string using nchar.
substring(my_vec, regexpr("__", my_vec) 2, nchar(my_vec))
Data
my_vec <- c("072:g__Caulobacter", "073:g__Romboutsia", "074:g__Blastocatella",
"076:c__Deltaproteobacteria", "077:g__Tatumella", "078:g__Fretibacterium")
CodePudding user response:
Does this work:
gsub('(\\d :[a-z]__)(.*)','\\2', vec)
[1] "Caulobacter" "Romboutsia" "Blastocatella" "Deltaproteobacteria" "Tatumella"
[6] "Fretibacterium"
CodePudding user response:
Another base R solution without needing a capture group is
my_vec <- c(
"072:g__Caulobacter",
"073:g__Romboutsia",
"074:g__Blastocatella",
"076:c__Deltaproteobacteria",
"077:g__Tatumella",
"078:g__Fretibacterium")
gsub("^. __", "", my_vec)
#[1] "Caulobacter" "Romboutsia" "Blastocatella"
#[4] "Deltaproteobacteria" "Tatumella" "Fretibacterium"
Explanation: "^. __" matches from the start of each string ("^") any character substring of length > 0 (". ") followed by a double underscore "__", and replaces this with an empty string "".
CodePudding user response:
I don't know R, but here would be a pure regex solution using two capturing groups: