I have a dataframe that looks like this:
> dput(df)
structure(list(V1 = c("Orthopedics", "Cardiac", "Interventional Radiology",
"Interventional Radiology", "Cardiac", "Cardiac", "Cardiac",
"Urology", "Cardiac", "Orthopedics", "Cardiac", "Neurosurgery",
"Cardiac", "Transplant", "Gynecology", "Cardiac", "Interventional Radiology",
"Interventional Radiology", "Cardiac", "Cardiac", "Urology",
"Vascular", "Neurosurgery", "Cardiac", "Cardiac", "Cardiac",
"Cardiac", "Cardiac", "General", "Cardiac", "Urology", "Cardiac",
"Interventional Radiology", "Neurosurgery", "Cardiac", "Cardiac",
"Urology", "Urology", "Thoracic", "Cardiac", "Neurosurgery",
"Urology", "Cardiac", "Urology", "Neurosurgery", "Neurosurgery",
"Orthopedics", "Cardiac", "Cardiac", "Orthopedics"), V2 = c("0",
"0", "0", "0", "0", "0", "0", "0", "1", "1", "0", "0", "0", "0",
"0", "1", "0", "1", "0", "0", "0", "0", "0", "0", "0", "0", "0",
"0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0",
"0", "0", "0", "1", "0", "0", "0", "0", "0", "0")), class = "data.frame", row.names = c(NA,
-50L))
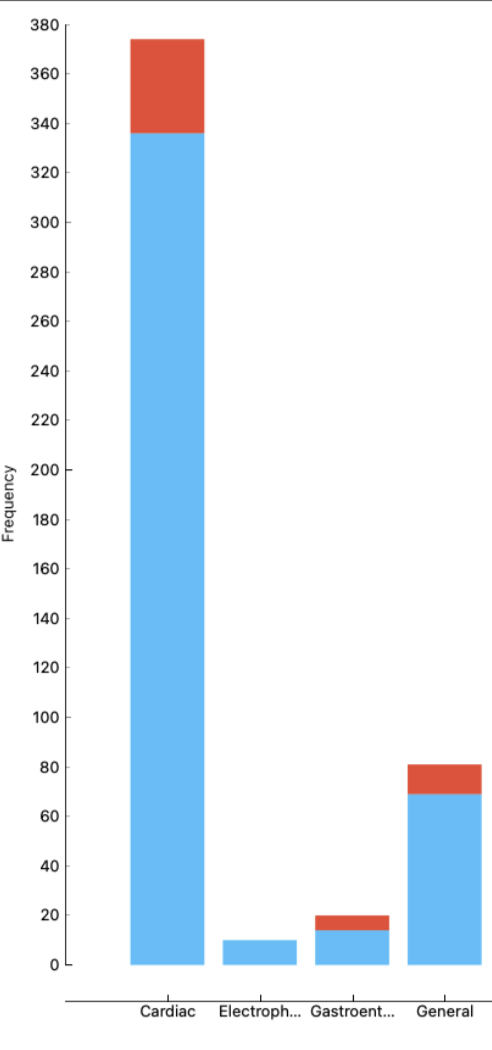
I want to create a stacked bar plot for each speciality (V1) that sums the number of 0's and 1's (shown in V2).
It should end up looking something like this, where the red represents the sum of 1's and the blue represents the sum of 0's.
CodePudding user response:
I think this is what you're after:
library(tidyverse)
df <- structure(list(V1 = c("Orthopedics", "Cardiac", "Interventional Radiology",
"Interventional Radiology", "Cardiac", "Cardiac", "Cardiac",
"Urology", "Cardiac", "Orthopedics", "Cardiac", "Neurosurgery",
"Cardiac", "Transplant", "Gynecology", "Cardiac", "Interventional Radiology",
"Interventional Radiology", "Cardiac", "Cardiac", "Urology",
"Vascular", "Neurosurgery", "Cardiac", "Cardiac", "Cardiac",
"Cardiac", "Cardiac", "General", "Cardiac", "Urology", "Cardiac",
"Interventional Radiology", "Neurosurgery", "Cardiac", "Cardiac",
"Urology", "Urology", "Thoracic", "Cardiac", "Neurosurgery",
"Urology", "Cardiac", "Urology", "Neurosurgery", "Neurosurgery",
"Orthopedics", "Cardiac", "Cardiac", "Orthopedics"), V2 = c("0",
"0", "0", "0", "0", "0", "0", "0", "1", "1", "0", "0", "0", "0",
"0", "1", "0", "1", "0", "0", "0", "0", "0", "0", "0", "0", "0",
"0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0",
"0", "0", "0", "1", "0", "0", "0", "0", "0", "0")), class = "data.frame", row.names = c(NA,
-50L))
df |>
group_by(V1, V2) |>
tally() |>
ggplot(aes(x = V1,
y = n,
fill = V2))
geom_col()
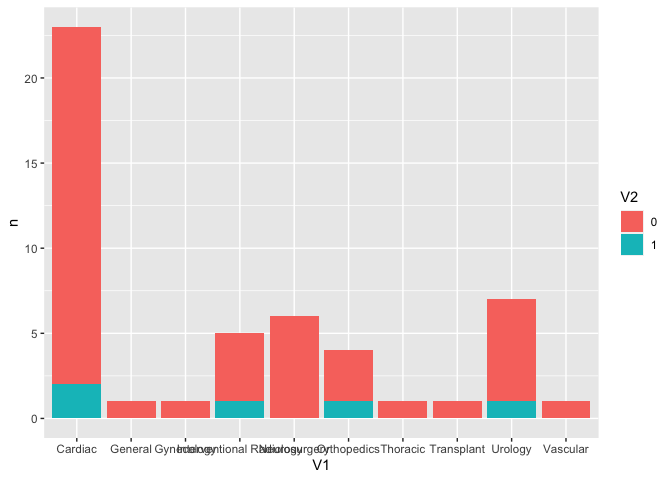
Created on 2022-09-27 by the reprex package (v2.0.1)