I have a dataset with 70 variables. The name of variables is like bio1 to bio70. I need to check the correlation of one variable, such as bio2, against the other 70 variables only. I used the following codes
## Generate scatterplot matrix
splom(MyData, panel = panel.smoothScatter, raster= TRUE, na=TRUE)
# Generate Correlations
cor(MyData, use = "pairwise.complete.obs")
corrplot.mixed(cor(MyData, use="pairwise.complete.obs"), lower.col = "black")
But these codes make a 70 by 70 matrix for me that I do not need it. How can I change these codes to give me the correlation matrix of one variable, such as bio2, against other variables? Thanks
CodePudding user response:
You didn't provide a dataset so I'll show you with R's iris dataset instead using both the tidyverse and correlation packages. First load the libraries:
#### Load Libraries ####
library(correlation)
library(tidyverse)
Then from there you can run a correlation matrix with the following code:
#### Correlation Matrix Default ####
iris %>%
correlation()
# Correlation Matrix (pearson-method)
Parameter1 | Parameter2 | r | 95% CI | t(148) | p
-------------------------------------------------------------------------
Sepal.Length | Sepal.Width | -0.12 | [-0.27, 0.04] | -1.44 | 0.152
Sepal.Length | Petal.Length | 0.87 | [ 0.83, 0.91] | 21.65 | < .001***
Sepal.Length | Petal.Width | 0.82 | [ 0.76, 0.86] | 17.30 | < .001***
Sepal.Width | Petal.Length | -0.43 | [-0.55, -0.29] | -5.77 | < .001***
Sepal.Width | Petal.Width | -0.37 | [-0.50, -0.22] | -4.79 | < .001***
Petal.Length | Petal.Width | 0.96 | [ 0.95, 0.97] | 43.39 | < .001***
p-value adjustment method: Holm (1979)
Observations: 150
If you want to select only the sepal variables, you can use this code instead:
#### Only Use Sepal Variables ####
iris %>%
select(Sepal.Length,
Sepal.Width) %>%
correlation()
Giving you this limited matrix now:
# Correlation Matrix (pearson-method)
Parameter1 | Parameter2 | r | 95% CI | t(148) | p
-------------------------------------------------------------------
Sepal.Length | Sepal.Width | -0.12 | [-0.27, 0.04] | -1.44 | 0.152
p-value adjustment method: Holm (1979)
Observations: 150
An alternative way of doing this is by deselecting the variables you dont want:
#### Alternative ####
iris %>%
select(-Petal.Length,
-Petal.Width) %>%
correlation()
Edit
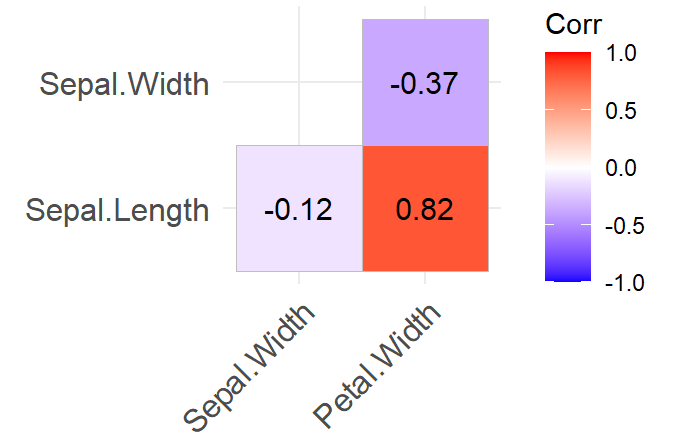
Seems you also wanted a correlation plot. I prefer using ggcorrplot cuz it looks better and its easier to work with. Here is a simple one only deselecting one variable from the matrix:
#### Using ggcorrplot ####
library(ggcorrplot)
corr <- iris %>%
select(-Petal.Length) %>%
correlation()
corr
ggcorrplot(corr = corr,
type = "lower",
lab = T)
Giving you this: