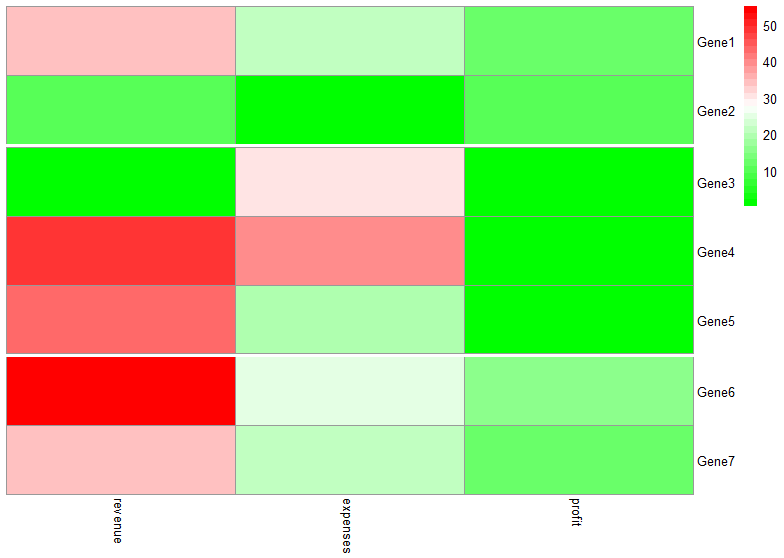
I have the following heatmap:
test <- data.frame(revenue=c(34, 10, 0.40, 49, 43, 55, 99),
expenses=c(22, 0.26, 31, 40, 20, 25, 22),
profit=c(12, 10, 0.14, 0.9, 0.8, 15, 16))
rownames(test) <- c("Gene1",
"Gene2",
"Gene3",
"Gene4",
"Gene5",
"Gene6",
"Gene7")
test
col_palette <- colorRampPalette(colors = c("green", "white", "red"))(30)
pheatmap(test,
cutree_rows = 1,
color = col_palette,
scale = "none",
cluster_cols = F,
cluster_rows=F)
and I just want to cut it horizontally so the final picture would look something like this:
I don't need cutting by clustering or whatsoever.
CodePudding user response:
Luckily, pheatmap has an argument to do exactly that: gaps_row. Try
pheatmap(test,
cutree_rows = 1,
color = col_palette,
scale = "none",
cluster_cols = F,
cluster_rows=F,
gaps_row=c(2,5))