I have a shiny app that outputs tables from a list of data frames using renderTable. A minimal example is shown below. I want the output table to have some rows output in scientific notation and the other rows output in percent format. How would I go about doing this?
library(shiny)
# create dummy CSVs -------------------------------------------------------
DF1 <- data.frame(x = c(1.1, 2.1, 3.4), y = c(5.6, 4.2, 6.7))
DF2 <- data.frame(x = c(2.4, 6.7, 8.9), y = c(4.1, 6.3, 8.4))
DF3 <- data.frame(x = c(3.5, 9.5, 3.2), y = c(1.4, 4.5, 7.2))
DF4 <- data.frame(x = c(2.1, 7.4, 8.2), y = c(1.5, 7.2, 6.2))
mapply(
write.csv,
x = list(DF1, DF2, DF3, DF4),
file = list("DF1.csv", "DF2.csv", "DF3.csv", "DF4.csv"),
row.names = FALSE
)
# shiny app ---------------------------------------------------------------
ui <- fluidPage(sidebarLayout(
sidebarPanel(
fileInput(
"files",
"Choose File",
multiple = TRUE,
accept = c(
"text/csv",
"text/comma-separated-values,text/plain",
".dp_txt",
".is_txt"
)
),
selectizeInput(
inputId = "selected_table",
label = "Table Selection",
choices = NULL,
selected = NULL,
multiple = FALSE
)
),
mainPanel(tableOutput("table"))
))
server <- function(input, output, session) {
observeEvent(input$files, {
freezeReactiveValue(input, "selected_table")
updateSelectizeInput(session,
inputId = "selected_table",
choices = input$files$name,
server = TRUE)
})
table_list <- reactive({
req(input$files)
setNames(lapply(input$files$datapath, function(x) {
fread(x)
}),
input$files$name)
})
output$table <- renderTable({
req(table_list(), input$selected_table)
table_list()[[input$selected_table]]
}, server = FALSE)
}
shinyApp(ui, server)
I know I can use the base R format() function to convert rows to scientific notation and can also use the label_percent() from the scales package to convert rows to percent format. I have not yet found a way to mix these format changes in the same table.
CodePudding user response:
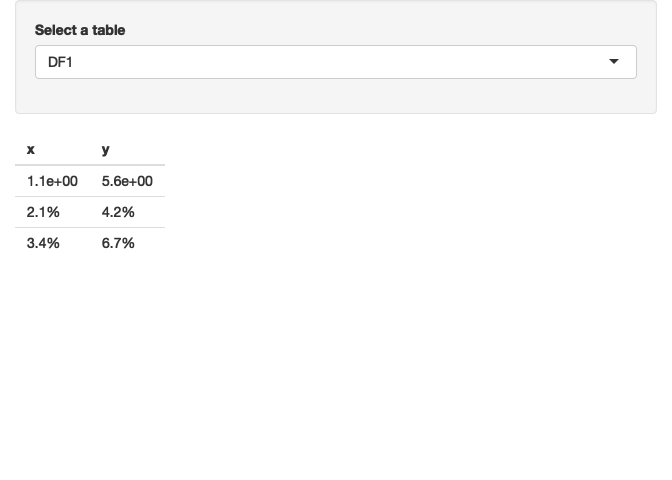
There are probably multiple approaches to achieve your desired result (and probably more efficient ones than my code below which uses lapply to loop over the columns and do the formatting of the rows. As you haven't specified which rows you want to format how I formatted row 1 in scientific and rows 2 to 3 in percent format:
Note: I dropped all the file input stuff to make the reprex a minimal reproducible example.
library(shiny)
DF1 <- data.frame(x = c(1.1, 2.1, 3.4), y = c(5.6, 4.2, 6.7))
DF2 <- data.frame(x = c(2.4, 6.7, 8.9), y = c(4.1, 6.3, 8.4))
DF3 <- data.frame(x = c(3.5, 9.5, 3.2), y = c(1.4, 4.5, 7.2))
DF4 <- data.frame(x = c(2.1, 7.4, 8.2), y = c(1.5, 7.2, 6.2))
ui <- fluidPage(sidebarLayout(
sidebarPanel(
selectInput("selected_table", "Select a table", paste0("DF", 1:4))
),
mainPanel(tableOutput("table"))
))
server <- function(input, output, session) {
table_list <- reactive({
setNames(list(DF1, DF2, DF3, DF4), paste0("DF", 1:4))
})
output$table <- renderTable({
req(table_list(), input$selected_table)
tbl <- table_list()[[input$selected_table]]
to_scientific <- 1
to_percent <- 2:3
lapply(tbl, function(x) {
xx <- x
xx[to_scientific] <- scales::scientific(x[to_scientific])
xx[to_percent] <- scales::percent(x[to_percent], scale = 1)
xx
})
}, server = FALSE)
}
shinyApp(ui, server)
#>
#> Listening on http://127.0.0.1:8112
Created on 2022-10-10 with reprex v2.0.2
