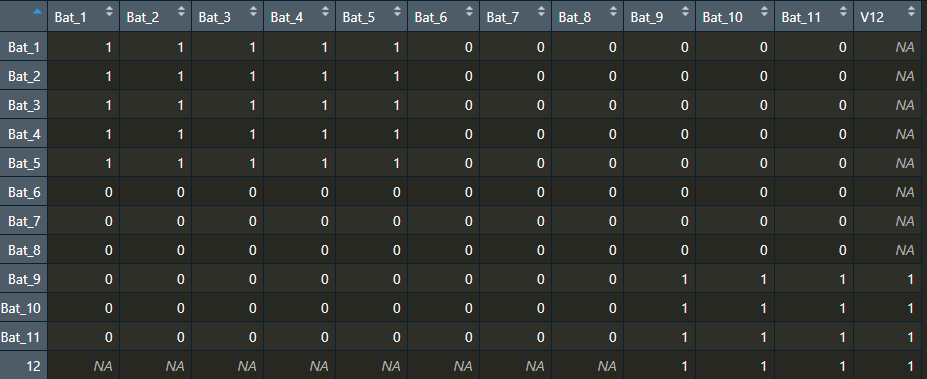
I have a matrix that will represent infection values of bats (can either be 1 or 0). The animals live in larger units ("roost", each roost is a submatrix), and the full matrix is the population. For starters, I am trying to fill my matrix with submatrices, values all equal to 1.
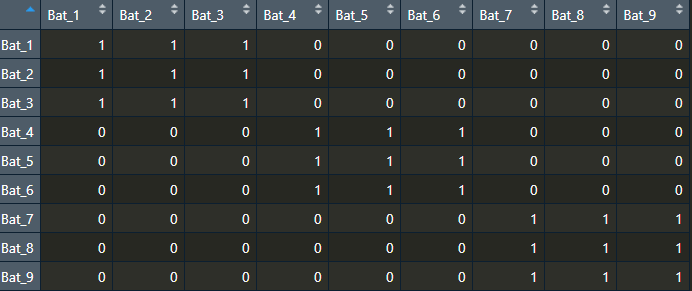
The code is currently working for roosts where number of bats is all the same. Ex:
# Define our variables
numRoosts = 3
# Uniformly sized roosts....
roostSizes = rep(3, numRoosts) # Each roost has 3 bats, looks like c(3, 3, 3)
# Adjaceny matrix describing connections between bats in all roosts
batAdjacencyMatrix <- as.data.frame(matrix(0, nrow = sum(roostSizes), ncol = sum(roostSizes)))
colnames(batAdjacencyMatrix) = rownames(batAdjacencyMatrix) = paste0("Bat_", 1:sum(roostSizes))
# Start filling in the network structure
n = 0
for(size in roostSizes){
# Fill the submatrices with dimension 'size x size' with 1's to create the subroost network
# Line below works for uniform roost sizes, but is buggy for nonuniform roost sizes
batAdjacencyMatrix[(1 n*size):((n 1)*size), (1 n*size):((n 1)*size)] <- 1 # Matrix indexing
# Increment the counter
n = n 1
}
This gives me the output I wanted:
The issue is when I try to change the roosts to nonuniform sizes:
numRoosts = 3
# If you want variable sized roosts....
minRoostPopulation = 2 # Min number of bats in a roost
maxRoostPopulation = 5 # Maximum number of bats in a roost
roostSizes <- round(runif(numRoosts, minRoostPopulation, maxRoostPopulation)) #Here c(5, 2, 4)
batAdjacencyMatrix <- as.data.frame(matrix(0, nrow = sum(roostSizes), ncol = sum(roostSizes)))
colnames(batAdjacencyMatrix) = rownames(batAdjacencyMatrix) = paste0("Bat_", 1:sum(roostSizes))
# Start filling in the network structure
n = 0
for(size in roostSizes){
# Fill the submatrices with dimension 'size x size' with 1's to create the subroost network
# Line below works for uniform roost sizes, but is buggy for nonuniform roost sizes
batAdjacencyMatrix[(1 n*size):((n 1)*size), (1 n*size):((n 1)*size)] <- 1 # Matrix indexing
# Increment the counter
n = n 1
}
There's something wrong with the indexing in my for loop- I can see when I put the numbers in manually. But I can't figure out how to define my submatrices so that the next number in the vector shifts down/right to the end of the matrix prior. Any thoughts? Thanks in advance!
CodePudding user response:
Your indexing is indeed somewhat off. If I were to modify your approach, I would do it like this:
numRoosts = 3
roostSizes <- c(5, 2, 4)
batAdjacencyMatrix <- as.data.frame(matrix(0, nrow = sum(roostSizes), ncol = sum(roostSizes)))
colnames(batAdjacencyMatrix) = rownames(batAdjacencyMatrix) = paste0("Bat_", 1:sum(roostSizes))
# Start filling in the network structure
n = 0
for(size in roostSizes){
# sum of sizes of preceding roosts
size.prior <- sum(head(roostSizes, n))
# indices of the current roost
ind <- size.prior (1:size)
batAdjacencyMatrix[ind, ind] <- 1 # Matrix indexing
# Increment the counter
n = n 1
}
However, an easier way is to use magic::adiag() which can build such block-diagonal matrices:
library(magic)
roostSizes <- c(5, 2, 4)
# create the three matrices of 1's
mat <- lapply(roostSizes, function(n) matrix(1, n, n))
# bind them diagonally
batAdjacencyMatrix <- do.call(adiag, mat)
rownames(batAdjacencyMatrix) <- colnames(batAdjacencyMatrix) <-
paste0('Bat_', seq_len(sum(roostSizes)))