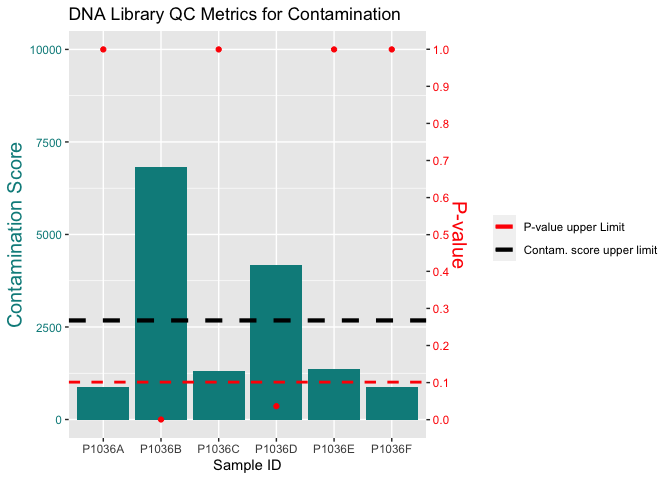
I very nearly have the plot that I wan't so I think this will be an easy fix for someone more R savvy than myself
Sample_ID <- c("P1014B", "P1014F", "P1036A", "P1036B", "P1036C", "P1036D", "P1036E", "P1036F")
`CONTAMINATION_SCORE (NA)` <- c(2677, 1021, 870, 6831, 1324, 4175, 1370, 875)
`CONTAMINATION_P_VALUE (NA)` <- c(0.101, 1.000, 1.000, 0.000, 1.000, 0.036, 1.000, 1.000)
contam_reads <- data.frame(Sample_ID, `CONTAMINATION_SCORE (NA)`, `CONTAMINATION_P_VALUE (NA)`, check.names = FALSE)
I have a plot that is very near to what I want:
cols = c("P-value upper Limit" = "black","Contam. score upper limit" = "red")
ggplot(contam_reads[-c(1,2),], aes(x=Sample_ID, y=`CONTAMINATION_SCORE (NA)`))
geom_bar(stat="identity", fill="cyan4")
geom_hline(aes(yintercept=contam_reads$`CONTAMINATION_SCORE (NA)`[1]), col = cols[2], size = 1.5, linetype=2)
geom_hline(aes(yintercept=contam_reads$`CONTAMINATION_P_VALUE (NA)`[1]*10000), col = cols[1], size = 1, linetype=2)
geom_point(aes(x=Sample_ID, y=`CONTAMINATION_P_VALUE (NA)`* 10000),stat="identity",color="red", size = 1.5, show.legend = T)
labs(title= "DNA Library QC Metrics for Contamination",
x="Sample ID",y="Contamination Score")
scale_y_continuous(sec.axis=sec_axis(~./10000,name="P-value", breaks = seq(0,1,0.1)))
theme(
axis.title.y = element_text(color = "cyan4",size=15),
axis.text.y = element_text(color = "cyan4"),
axis.title.y.right = element_text(color = "red"),
axis.text.y.right = element_text(color = "red")
)
scale_colour_manual(values=cols)
I can't get the legend to be a line and not dots, and I can't get the first dot in the legend to turn black.
CodePudding user response:
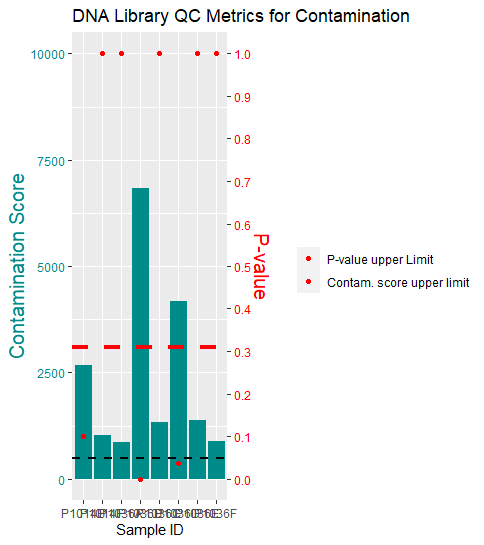
You could fix your legend by mapping on aesthetics, i.e. instead of setting colors for the hlines as arguments map on the color aes and set show.legend=FALSE for geom_point to show just the lines. To this end I also use the data argument to pass just the first line of the dataset to the hlines.
Note 1: I added a named vector with labels for your scores and values. Also as names I use abbreviations for both the labels and color vector which makes it much easier to refer to a specific value in the ggplot code and makes the code easier to read.
Note 2: In my plot the colors for the lines are swapped which is right if p-values should be "red" and scores "black". Perhaps you confused that which could easily happen if one uses e.g. cols[1] to refer to colors.
cols <- c("pval" = "red", "score" = "black")
labels <- c("pval" = "P-value upper Limit", "score" = "Contam. score upper limit")
library(ggplot2)
ggplot(contam_reads[-c(1, 2), ], aes(x = Sample_ID, y = `CONTAMINATION_SCORE (NA)`))
geom_bar(stat = "identity", fill = "cyan4")
geom_hline(
data = contam_reads[1, ],
aes(yintercept = `CONTAMINATION_SCORE (NA)`, color = "score"),
size = 1.5, linetype = 2
)
geom_hline(
data = contam_reads[1, ],
aes(yintercept = `CONTAMINATION_P_VALUE (NA)` * 10000, color = "pval"),
size = 1, linetype = 2
)
geom_point(aes(x = Sample_ID, y = `CONTAMINATION_P_VALUE (NA)` * 10000, color = "pval"),
stat = "identity", size = 1.5, show.legend = FALSE
)
labs(
title = "DNA Library QC Metrics for Contamination",
x = "Sample ID", y = "Contamination Score",
color = NULL
)
scale_y_continuous(sec.axis = sec_axis(~ . / 10000, name = "P-value", breaks = seq(0, 1, 0.1)))
theme(
axis.title.y = element_text(color = "cyan4", size = 15),
axis.text.y = element_text(color = "cyan4"),
axis.title.y.right = element_text(color = "red"),
axis.text.y.right = element_text(color = "red")
)
scale_colour_manual(values = cols, labels = labels)