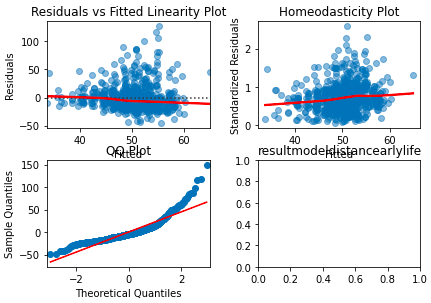
I have the following code:
listofmodels = [resultmodeldistancearlylife,resultmodeldurationearlylife,resultmodeldistancenavigate,resultmodeldurationnavigate,resultmodeldistancetravel,resultmodeldurationtravel,resultmodeldistancevideo,resultmodeldurationvideo,resultmodeldistancelifestyle,resultmodeldurationlifestyle,resultmodeldistancegrow,resultmodeldurationgrow,resultmodeldistancesleep,resultmodeldurationsleep,resultmodeldistancedemographics,resultmodeldurationdemographics]
names = ['resultmodeldistancearlylife','resultmodeldurationearlylife','resultmodeldistancenavigate','resultmodeldurationnavigate','resultmodeldistancetravel','resultmodeldurationtravel','resultmodeldistancevideo','resultmodeldurationvideo','resultmodeldistancelifestyle','resultmodeldurationlifestyle','resultmodeldistancegrow','resultmodeldurationgrow','resultmodeldistancesleep','resultmodeldurationsleep','resultmodeldistancedemographics','resultmodeldurationdemographics']
for i in range(len(listofmodels)):
fig, axes = plt.subplots(nrows=2, ncols=2)
plt.tight_layout()
plt.title(names[i],loc='left')
sns.residplot(listofmodels[i].predict(), y, lowess=True, scatter_kws={'alpha': 0.5}, line_kws={'color':'red'}, ax=axes[0,0])
axes[0,0].title.set_text('Residuals vs Fitted Linearity Plot')
axes[0,0].set(xlabel='Fitted', ylabel='Residuals')
sm.ProbPlot(listofmodels[i].resid).qqplot(line='s', color='#1f77b4', ax=axes[1,0])
axes[1,0].title.set_text('QQ Plot')
standardized_resid1 = np.sqrt(np.abs(listofmodels[i].get_influence().resid_studentized_internal))
sns.regplot(listofmodels[i].predict(), standardized_resid1, color='#1f77b4', lowess=True, scatter_kws={'alpha': 0.5}, line_kws={'color':'red'}, ax=axes[0,1])
axes[0,1].title.set_text('Homeodasticity Plot')
axes[0,1].set(xlabel='Fitted', ylabel='Standardized Residuals')
However, when I run the code, the subplot graph axis labels overlap:
The plt.tight_layout() function thus does not seem to be working.
Would anybody be able to give me a helping hand? I would be so grateful
CodePudding user response:
This was solved by using the following command:
fit.tight_layout(pad=3.2)
I also changed the position of the figure label using:
plt.title(names[i],x=-1,y=3)
Final code:
listofmodels = [resultmodeldistancearlylife,resultmodeldurationearlylife,resultmodeldistancenavigate,resultmodeldurationnavigate,resultmodeldistancetravel,resultmodeldurationtravel,resultmodeldistancevideo,resultmodeldurationvideo,resultmodeldistancelifestyle,resultmodeldurationlifestyle,resultmodeldistancegrow,resultmodeldurationgrow,resultmodeldistancesleep,resultmodeldurationsleep,resultmodeldistancedemographics,resultmodeldurationdemographics]
names = ['resultmodeldistancearlylife','resultmodeldurationearlylife','resultmodeldistancenavigate','resultmodeldurationnavigate','resultmodeldistancetravel','resultmodeldurationtravel','resultmodeldistancevideo','resultmodeldurationvideo','resultmodeldistancelifestyle','resultmodeldurationlifestyle','resultmodeldistancegrow','resultmodeldurationgrow','resultmodeldistancesleep','resultmodeldurationsleep','resultmodeldistancedemographics','resultmodeldurationdemographics']
for i in range(len(listofmodels)):
fig, axes = plt.subplots(nrows=2, ncols=2)
fig.tight_layout(pad=3.2)
plt.title(names[i],x=-1,y=3)
sns.residplot(listofmodels[i].predict(), y, lowess=True, scatter_kws={'alpha': 0.5}, line_kws={'color':'red'}, ax=axes[0,0])
axes[0,0].set_title('Residuals vs Fitted Linearity Plot',y=1.05)
axes[0,0].set(xlabel='Fitted', ylabel='Residuals')
sm.ProbPlot(listofmodels[i].resid).qqplot(line='s', color='#1f77b4', ax=axes[1,0])
axes[1,0].set_title('QQ Plot',y=1.05)
standardized_resid1 = np.sqrt(np.abs(listofmodels[i].get_influence().resid_studentized_internal))
sns.regplot(listofmodels[i].predict(), standardized_resid1, color='#1f77b4', lowess=True, scatter_kws={'alpha': 0.5}, line_kws={'color':'red'}, ax=axes[0,1])
axes[0,1].set_title('Homeodasticity Plot',y=1.05)
axes[0,1].set(xlabel='Fitted', ylabel='Standardized Residuals')
CodePudding user response:
The title and labels, and maybe axes numbers, are placed in the space between the grids.
The default space is just enough for one text.
You have to increase the spacing with figure.add_gridspec()
You can also move the titles and labels outside the for loop.
listofmodels = [resultmodeldistancearlylife,resultmodeldurationearlylife,resultmodeldistancenavigate,resultmodeldurationnavigate,resultmodeldistancetravel,
resultmodeldurationtravel,resultmodeldistancevideo,resultmodeldurationvideo,resultmodeldistancelifestyle,resultmodeldurationlifestyle,
resultmodeldistancegrow,resultmodeldurationgrow,resultmodeldistancesleep,resultmodeldurationsleep,resultmodeldistancedemographics,resultmodeldurationdemographics]
names = ['resultmodeldistancearlylife','resultmodeldurationearlylife','resultmodeldistancenavigate','resultmodeldurationnavigate','resultmodeldistancetravel',
'resultmodeldurationtravel','resultmodeldistancevideo','resultmodeldurationvideo','resultmodeldistancelifestyle','resultmodeldurationlifestyle',
'resultmodeldistancegrow','resultmodeldurationgrow','resultmodeldistancesleep','resultmodeldurationsleep','resultmodeldistancedemographics','resultmodeldurationdemographics']
fig = plt.figure()
gs = fig.add_gridspec(2, 2, hspace=0.7, wspace=0.7)
axes = gs.subplots()
# plt.tight_layout()
axes[0,0].title.set_text('Residuals vs Fitted Linearity Plot')
axes[0,0].set(xlabel='Fitted', ylabel='Residuals')
axes[1,0].title.set_text('QQ Plot')
axes[0,1].title.set_text('Homeodasticity Plot')
axes[0,1].set(xlabel='Fitted', ylabel='Standardized Residuals')
for i in range(len(listofmodels)):
axes[1,1].title(names[i],loc='left')
sns.residplot(listofmodels[i].predict(), y, lowess=True, scatter_kws={'alpha': 0.5}, line_kws={'color':'red'}, ax=axes[0,0])
sm.ProbPlot(listofmodels[i].resid).qqplot(line='s', color='#1f77b4', ax=axes[1,0])
standardized_resid1 = np.sqrt(np.abs(listofmodels[i].get_influence().resid_studentized_internal))
sns.regplot(listofmodels[i].predict(), standardized_resid1, color='#1f77b4', lowess=True, scatter_kws={'alpha': 0.5}, line_kws={'color':'red'}, ax=axes[0,1])