I have a pipe delimited files with header and body. Header data has fewer columns than the body. In data flow, I am splitting header and body data and doing transformations on them and union transformation is applied to transformed header and body data. While joining both the data, I am getting additional pipes at the end of the header.
Source data will be like
Header1|id1
Header2|id2
1|Debashish|1500|30
2|Susmitha|1900|20
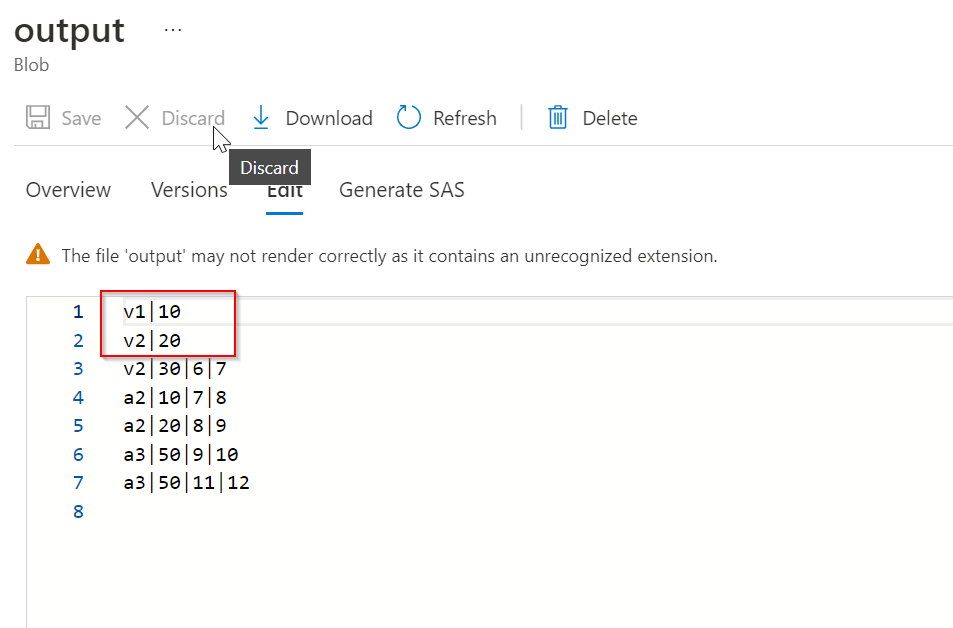
After doing transformations,
Header1|id1||
Header2|id2||
I need to remove the extra pipelines at the end of header, It should be same as source. How can I do this.
CodePudding user response:
Before copying the data from union transformation to the sink, you can concatenate all columns into a single column separated by pipe symbol and then trim the extra pipes available in the end of the data using derived column activity. I tried to repro this in my environment. Below are the detailed steps.
- A sample source file is taken as in below image.
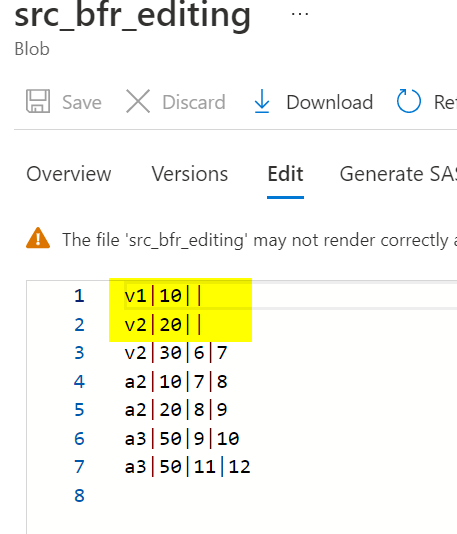
- Source transformation is added with source dataset in dataflow activity.
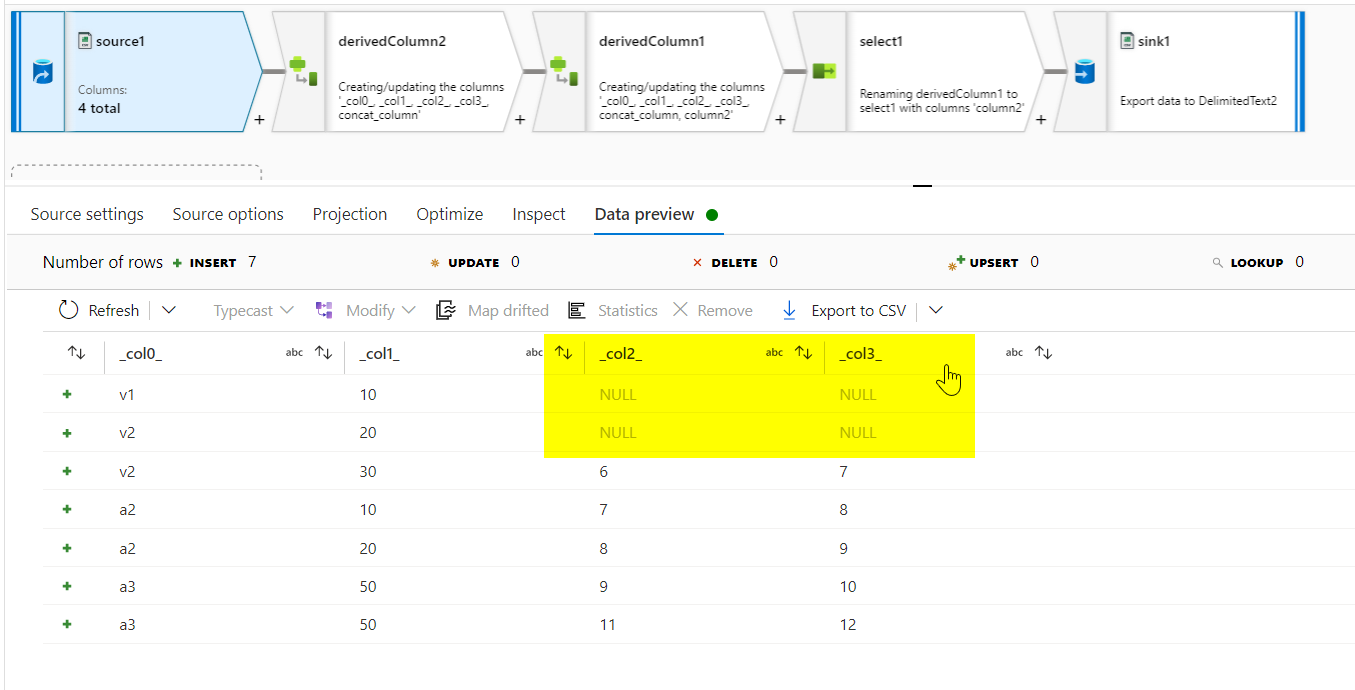
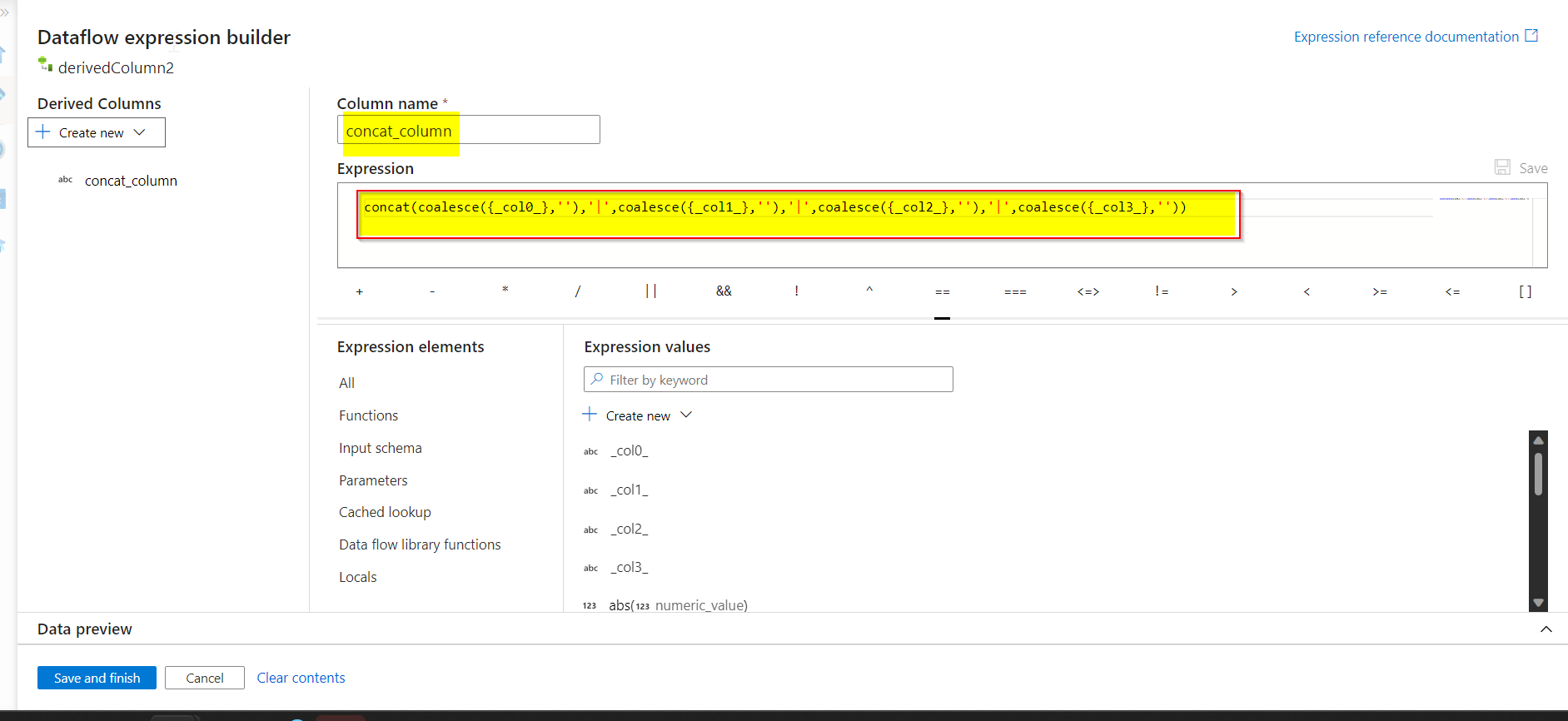
- Then, a new column called
concat_columnis created using derivedcolumn transformation. Value for the column is given asconcat(coalesce({_col0_},''),'|',coalesce({_col1_},''),'|',coalesce({_col2_},''),'|',coalesce({_col3_},''))
- Then again, a derived column transformation is added to remove the pipes from the data. Column2 is created and value is given as
rtrim(concat_column,'||').
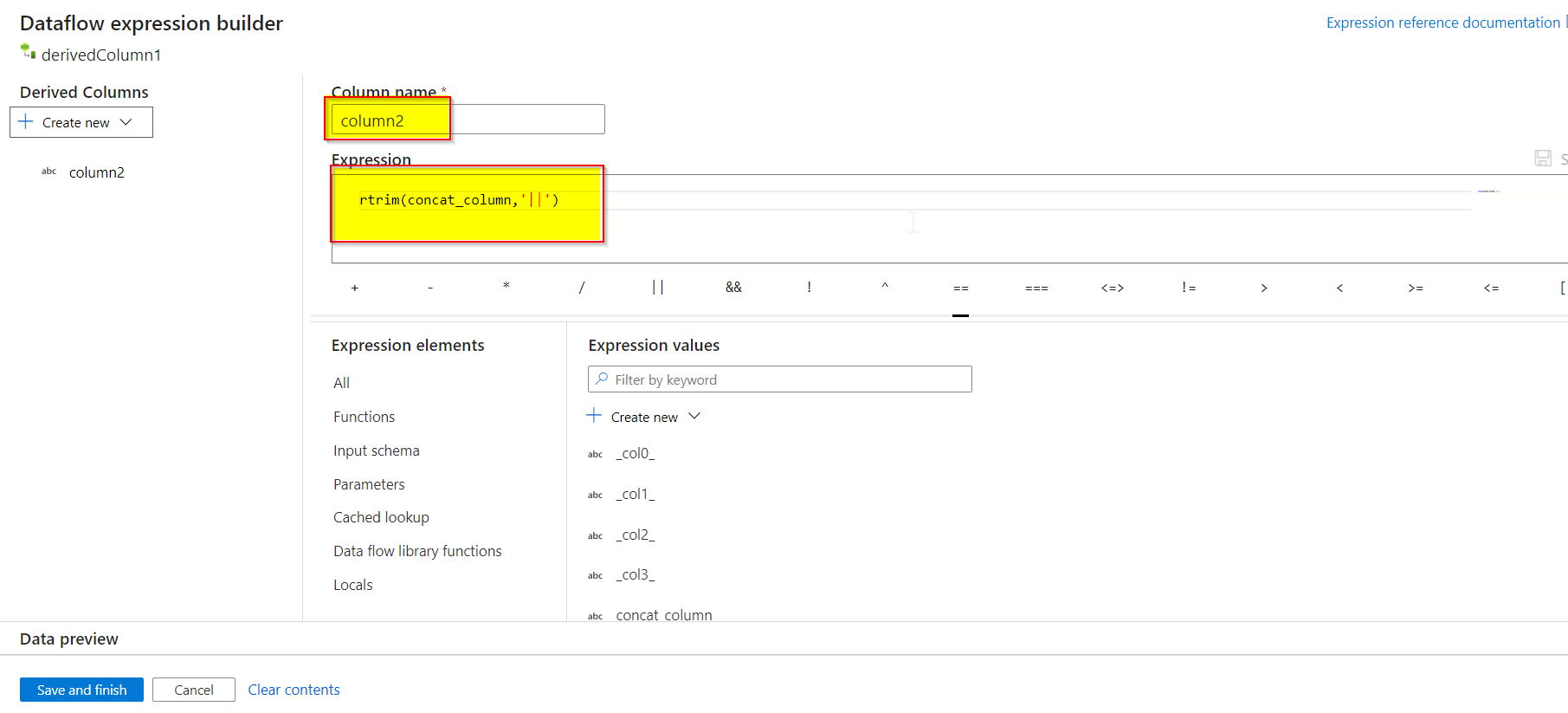
Results of concat_column and column2 :
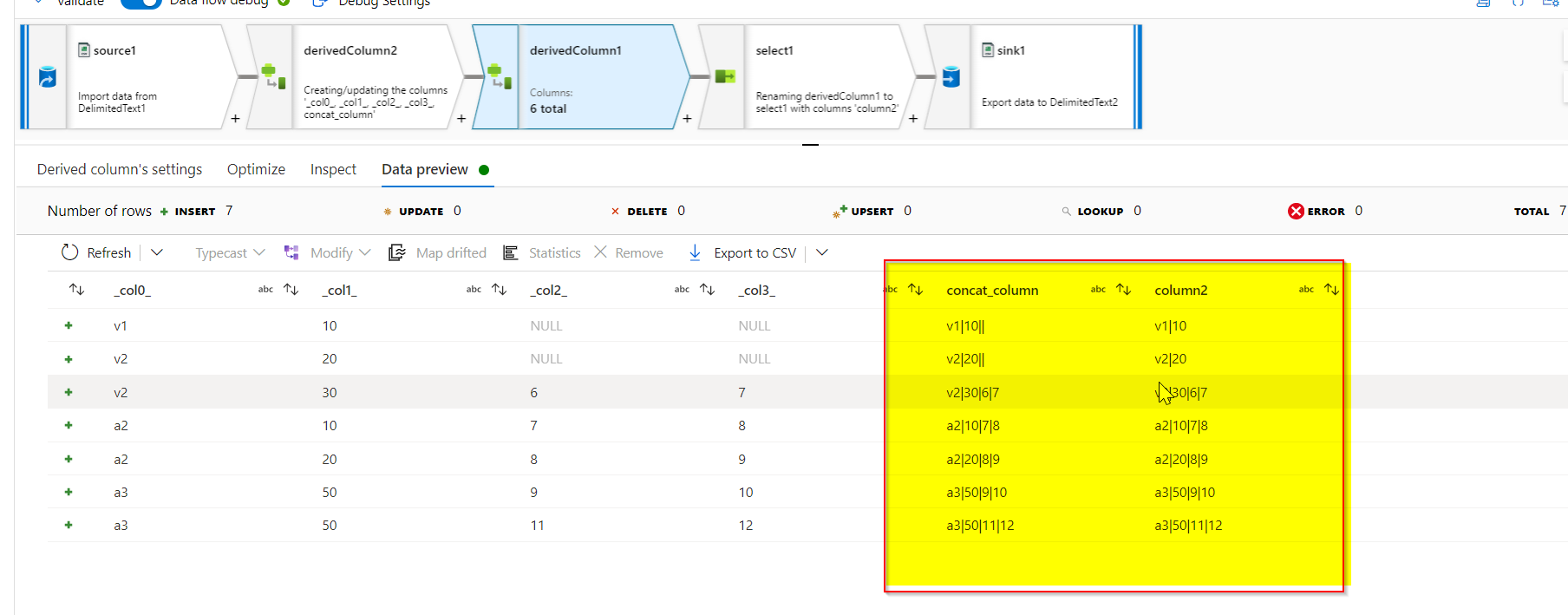
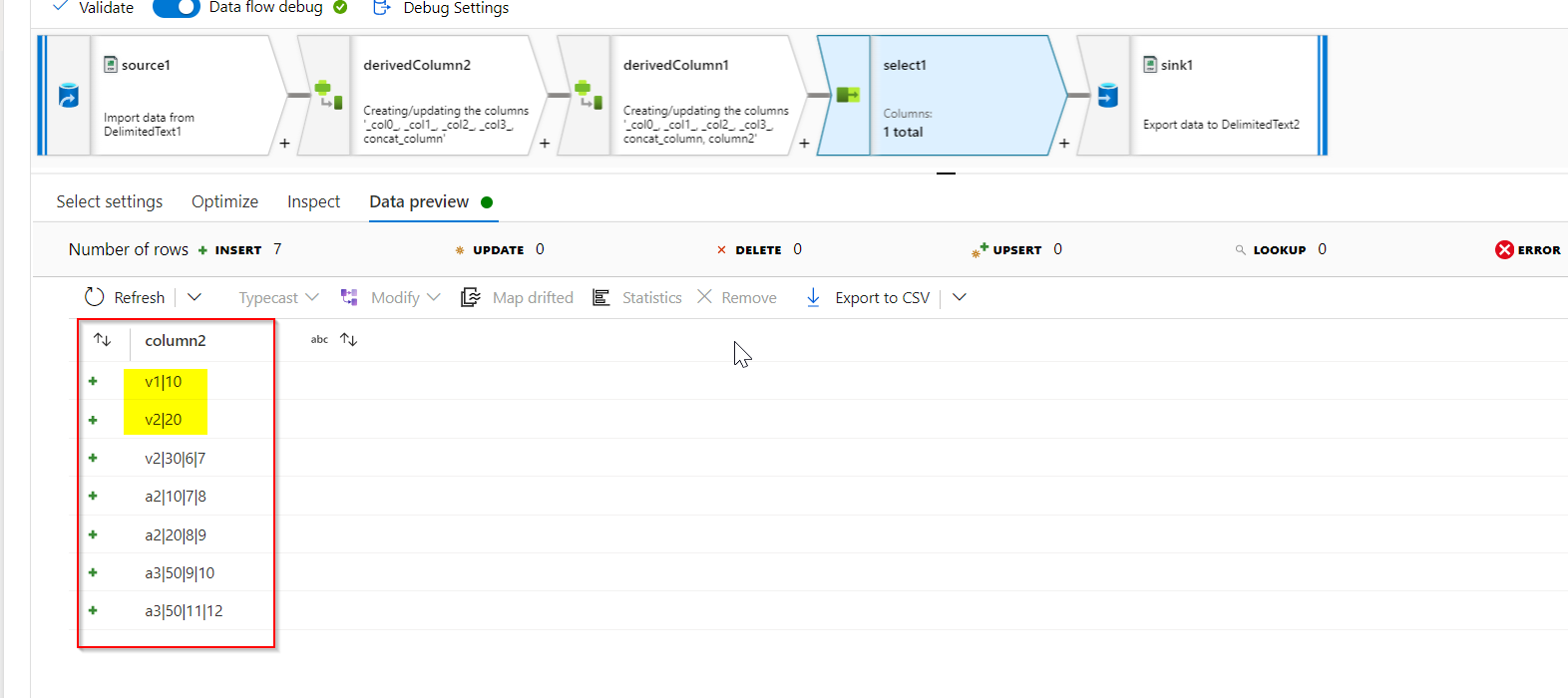
Select transformation is added only to select the column2
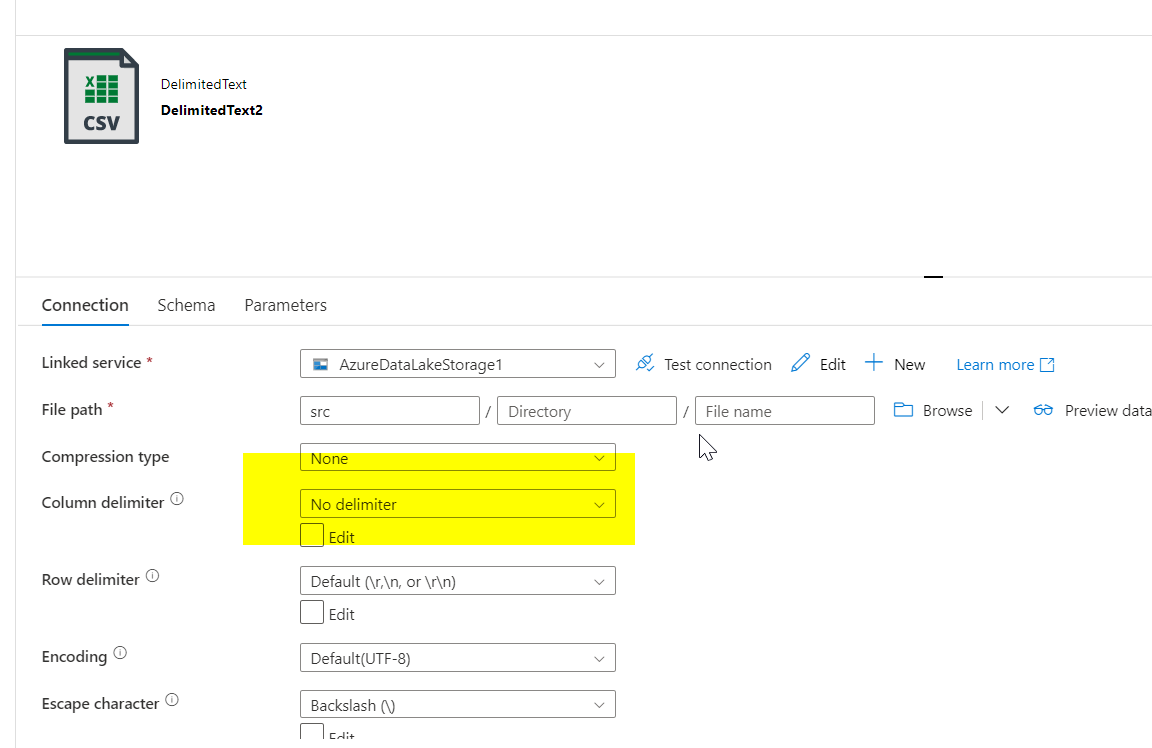
Sink transformation is added and for sink dataset, different delimiter is given (other than pipe symbol. Here no delimiter is used).
- After executing the pipeline, Output file has no pipe symbol at the end.