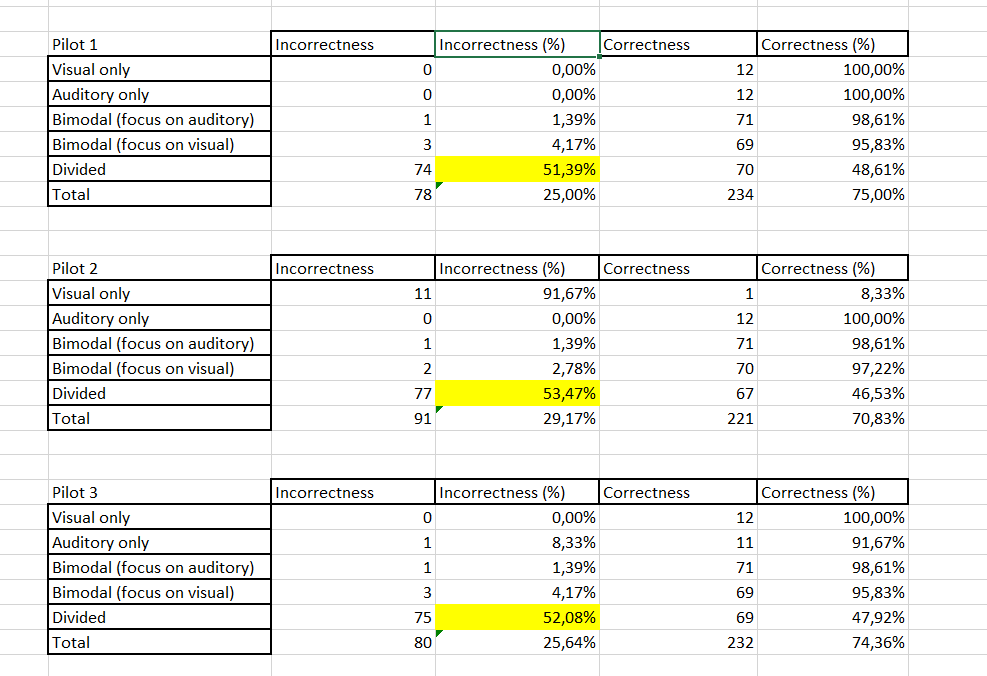
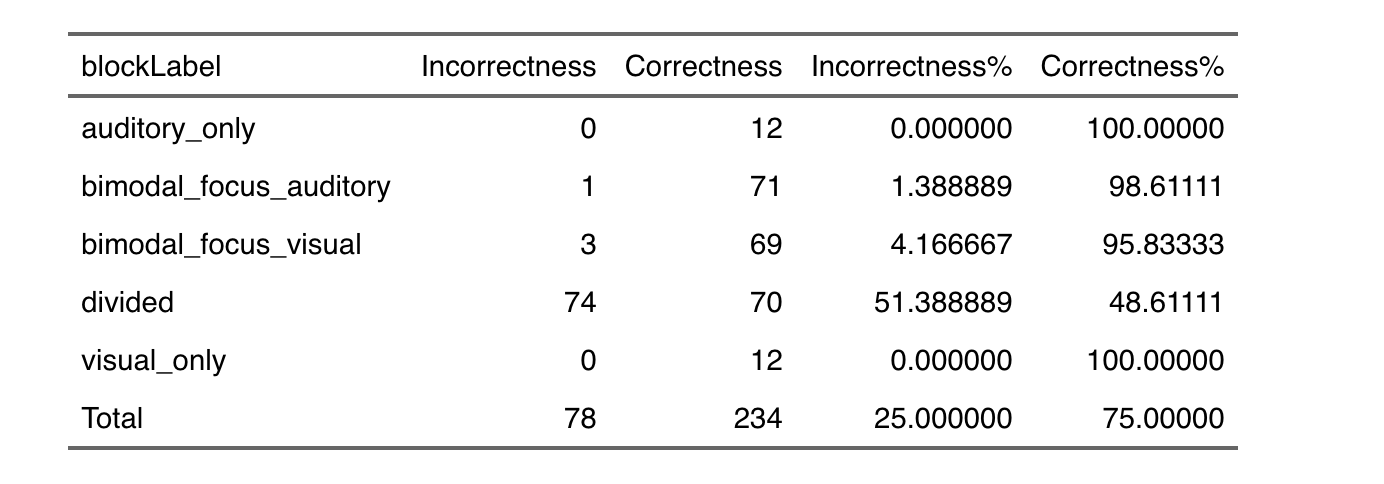
I have the following tibble
structure(list(blockLabel = structure(c(1L, 2L, 3L, 4L, 5L, 1L,
2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 1L, 2L,
3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L), .Label = c("auditory_only",
"bimodal_focus_auditory", "bimodal_focus_visual", "divided",
"visual_only"), class = "factor"), trial_resp.corr = structure(c(1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L), .Label = c("0",
"1"), class = "factor"), participant = structure(c(1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L), .Label = c("pilot01",
"pilot02", "pilot03"), class = "factor"), Freq = c(0L, 1L, 3L,
74L, 0L, 12L, 71L, 69L, 70L, 12L, 0L, 1L, 2L, 77L, 11L, 12L,
71L, 70L, 67L, 1L, 1L, 1L, 3L, 75L, 0L, 11L, 71L, 69L, 69L, 12L
), tc = c(12, 72, 72, 144, 12, 12, 72, 72, 144, 12, 12, 72, 72,
144, 12, 12, 72, 72, 144, 12, 12, 72, 72, 144, 12, 12, 72, 72,
144, 12), freq = c(0, 1.38888888888889, 4.16666666666667, 51.3888888888889,
0, 100, 98.6111111111111, 95.8333333333333, 48.6111111111111,
100, 0, 1.38888888888889, 2.77777777777778, 53.4722222222222,
91.6666666666667, 100, 98.6111111111111, 97.2222222222222, 46.5277777777778,
8.33333333333333, 8.33333333333333, 1.38888888888889, 4.16666666666667,
52.0833333333333, 0, 91.6666666666667, 98.6111111111111, 95.8333333333333,
47.9166666666667, 100)), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -30L))
I would like to create three different tables according to the levels of participant variable. More or less the final result should be like the following one:
I have started scripting the following code (since I am looking for to do this via a dplyr, apply family, loops or map function)
list %>% as_data_frame() %>%
select(blockLabel, trial_resp.corr, participant, Freq, freq) %>%
map(~ flextable(.x))
But unfortunately, I get the following error code:
Error in flextable(.x) : is.data.frame(data) is not TRUE
I am not an expert in this method thus if you would have something to suggest about fixing the problem and sharing knowledge to achieve the final result, please just let me know (in passing I specify that Correctness corresponds to 1 and Incorrcetness to 0 of trial_resp.corr variable)
Thanks
CodePudding user response:
Here is one method where we modify the 'trial_resp.corr' values to 'Incorrectness', 'Correctness' based on 0, 1 values, then select the columns of interest, split by the 'participant' column to create a list of datasets, loop over the list with map, reshape data from long to wide with pivot_wider and then convert to flextable
library(dplyr)
library(tidyr)
library(purrr)
library(flextable)
library(stringr)
library(janitor)
lst1 <- df1 %>%
mutate( trial_resp.corr = recode(trial_resp.corr,
'0'= 'Incorrectness', '1' = "Correctness")) %>%
select(blockLabel, trial_resp.corr, participant, Freq, `Freq%` = freq) %>%
split(.$participant) %>%
map(~ .x %>%
select(-participant) %>%
pivot_wider(names_from = trial_resp.corr,
values_from = c(Freq, `Freq%`), values_fill = 0,
names_glue ="{trial_resp.corr}{str_remove(.value, 'Freq' )}") %>%
adorn_totals() %>%
mutate(across(ends_with("%"),
~ replace(.x, n(), 100 *get(str_remove(cur_column(),
fixed("%")))[n()]/sum(Incorrectness[n()], Correctness[n()])))) %>%
flextable)
If we want to create objects in the global env
list2env(lst1, .GlobalEnv)
-output
lst1[[1]]
CodePudding user response:
I would propose the following (for dplyr):
library(dplyr)
dfs <- list()
for (prt in levels(df$participant)){
dfs[[prt]] <- df %>% filter(participant == prt) %>% select(-participant)
}
You get the three tables for any required further manipulations:
> dfs
$pilot01
# A tibble: 10 × 5
blockLabel trial_resp.corr Freq tc freq
<fct> <fct> <int> <dbl> <dbl>
1 auditory_only 0 0 12 0
2 bimodal_focus_auditory 0 1 72 1.39
3 bimodal_focus_visual 0 3 72 4.17
4 divided 0 74 144 51.4
5 visual_only 0 0 12 0
6 auditory_only 1 12 12 100
7 bimodal_focus_auditory 1 71 72 98.6
8 bimodal_focus_visual 1 69 72 95.8
9 divided 1 70 144 48.6
10 visual_only 1 12 12 100
$pilot02
# A tibble: 10 × 5
blockLabel trial_resp.corr Freq tc freq
<fct> <fct> <int> <dbl> <dbl>
1 auditory_only 0 0 12 0
2 bimodal_focus_auditory 0 1 72 1.39
3 bimodal_focus_visual 0 2 72 2.78
4 divided 0 77 144 53.5
5 visual_only 0 11 12 91.7
6 auditory_only 1 12 12 100
7 bimodal_focus_auditory 1 71 72 98.6
8 bimodal_focus_visual 1 70 72 97.2
9 divided 1 67 144 46.5
10 visual_only 1 1 12 8.33
$pilot03
# A tibble: 10 × 5
blockLabel trial_resp.corr Freq tc freq
<fct> <fct> <int> <dbl> <dbl>
1 auditory_only 0 1 12 8.33
2 bimodal_focus_auditory 0 1 72 1.39
3 bimodal_focus_visual 0 3 72 4.17
4 divided 0 75 144 52.1
5 visual_only 0 0 12 0
6 auditory_only 1 11 12 91.7
7 bimodal_focus_auditory 1 71 72 98.6
8 bimodal_focus_visual 1 69 72 95.8
9 divided 1 69 144 47.9
10 visual_only 1 12 12 100