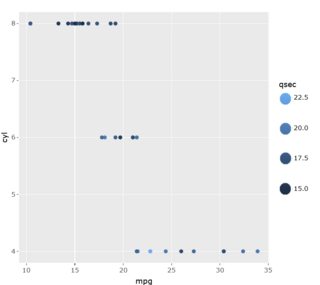
Package shinyscreenshot is not able to print plotly colorbars (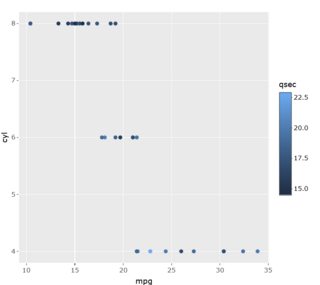
Goal
It doesn't mattert if there are 4, 5 or X datapoints in legend.
MWE
library(ggplot2)
library(plotly)
ggplotly(
ggplot(data=mtcars,
aes(x=mpg, y=cyl, color=qsec))
geom_point()
)
CodePudding user response:
This is only a partial answer; you can change the shape and text of the legend while maintaining its gradient by using a combination of scale_color_continuous and guides(color = guide_legend()), but this will only show up as a ggplot object. For some reason, the legend disappears when you add the plot to ggplotly(), the legend disappears. I suspect that arguments specific to the legend 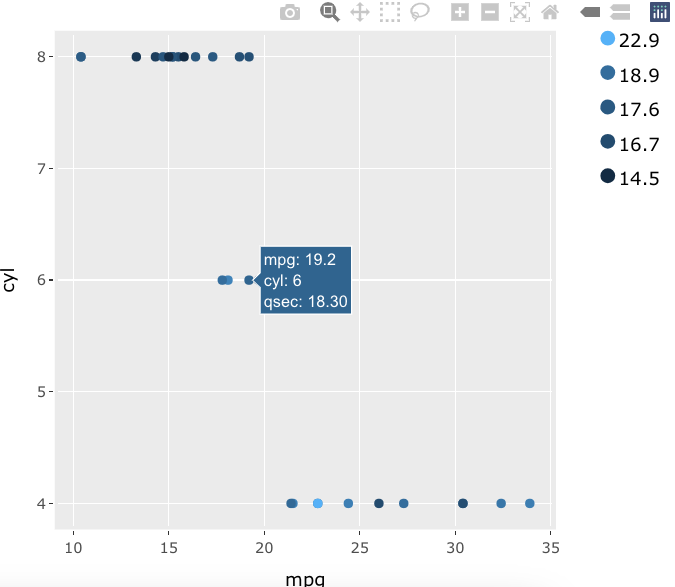
All of that code in one chunk:
library(tidyverse)
library(plotly)
# original plot
plt <- ggplotly(
ggplot(data=mtcars,
aes(x=mpg, y=cyl, color=qsec))
geom_point()
)
g <- ggplot(data=mtcars,
aes(x=mpg, y=cyl, color=qsec))
geom_point()
# color by qsec values frame
colByVal <- cbind(ggplot_build(g)$data[[1]], mtcars) %>%
as.data.frame() %>%
select(colour, qsec) %>% arrange(qsec) %>%
group_by(colour) %>%
summarise(qsec = median(qsec)) %>% as.data.frame()
parts <- summary(colByVal$qsec)
# drop the mean or median (the same color probably)
parts <- parts[-4]
vals <- lapply(parts, function(k) {
DescTools::Closest(colByVal$qsec, k)[1]
}) %>% unlist(use.names = F)
cols <- colByVal %>%
filter(qsec %in% vals) %>% select(colour) %>%
unlist(use.names = F)
ys <- seq(from = .7, by = .07, length.out = length(cols))
# create shapes
shp <- function(y, cr) { # y0, and fillcolor
list(type = "circle",
xref = "paper", x0 = 1.1, x1 = 1.125,
yref = "paper", y0 = y, y1 = y .025,
fillcolor = cr, yanchor = "center",
line = list(color = cr))
}
# create labels
ano <- function(ya, lab) { # y and label
list(x = 1.13, y = ya .035, text = lab,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = 'top',
showarrow = F)
}
# the shapes list
shps <- lapply(1:length(cols),
function(j) {
shp(ys[j], cols[j])
})
# the labels list
labs <- lapply(1:length(cols),
function(i) {
ano(ys[i], as.character(vals[i]))
})
# ggplot > ggplotly adds an empty shape; this conflicts with calling it in
# layout(); we'll replace 'shapes' first
plt$x$layout$shapes <- shps
plt %>% hide_colorbar() %>%
layout(annotations = labs, showlegend = F,
margin = list(t = 30, r = 100, l = 50, b = 30, pad = 3))