I have the following function below
I'd like to sample from this CDF (x values) but can't find the inverse function by hand. Is there a way to do it in R?
a <- 3.1
be <- -0.15
ga <- 0.78
delt <- 0.12
c<- 3.5
b <- exp(a be*c*(1 c))
g <- exp((ga delt*c)*c)
flc_F <- function(x){
#x between 0 and 1
if (x<1){
return ((b*(g-1)*(1-b^x))/(b*(g-1) (1-b*g)*b^x))
} else {
return (1)
}
}
CodePudding user response:
You can create an inverse with uniroot:
a <- 3.1
be <- -0.15
ga <- 0.78
delt <- 0.12
c<- 3.5
b <- exp(a be*c*(1 c))
g <- exp((ga delt*c)*c)
flc_F <- function(x){
#x between 0 and 1
if (x<1){
return ((b*(g-1)*(1-b^x))/(b*(g-1) (1-b*g)*b^x))
} else {
return (1)
}
}
flc_F <- Vectorize(flc_F)
# Quantile function
Qflc_F <- function(p){
sapply(p, function(.x) uniroot(
function(p) flc_F(p) - .x,
interval = c(0, 1e16)
)[["root"]])
}
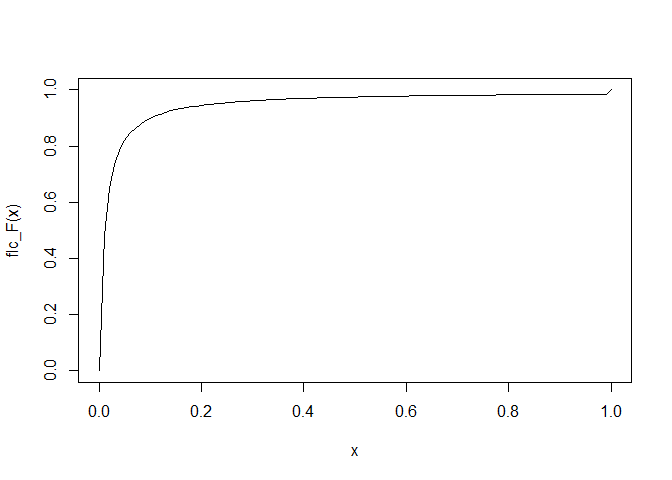
Plot of the functions. CDF...
curve(flc_F(x), from = 0, to = 1)
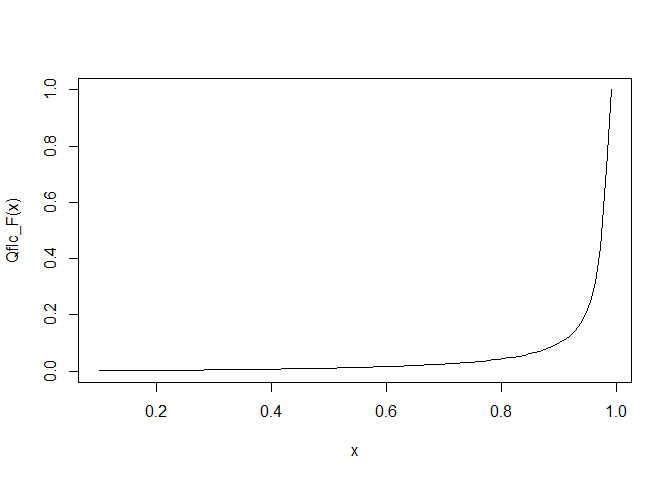
And Inverse (that is, Quantile function:
curve(Qflc_F(x), from = 0.1, to = 0.99)
PD: Note two things:
1 - I vectorized your original function using Vectorize(), thus you can use it with vectors (there are possibly more efficient ways though)
2 - I set a limit for search the root in uniroot between 0 and 1e16, of course that function could be refined to treat Inf values in better way.
CodePudding user response:
Since you said you have the cdf, You can use the inverse cdf method to sample from the density:
a <- 3.1
be <- -0.15
ga <- 0.78
delt <- 0.12
c<- 3.5
b <- exp(a be*c*(1 c))
g <- exp((ga delt*c)*c)
flc_F <- function(x){
#x between 0 and 1
if(x<0) return(NA)
if (x<1){
return ((b*(g-1)*(1-b^x))/(b*(g-1) (1-b*g)*b^x))
} else {
return (1)
}
}
# Inverse of the cdf:
flc_Inv <- function(x)optimise(\(y)(flc_F(y)-x)^2,c(0,1) )[[1]]
Now just sample points from uniform(0,1) and invert them to follow the said distribution using the inverse cdf function:
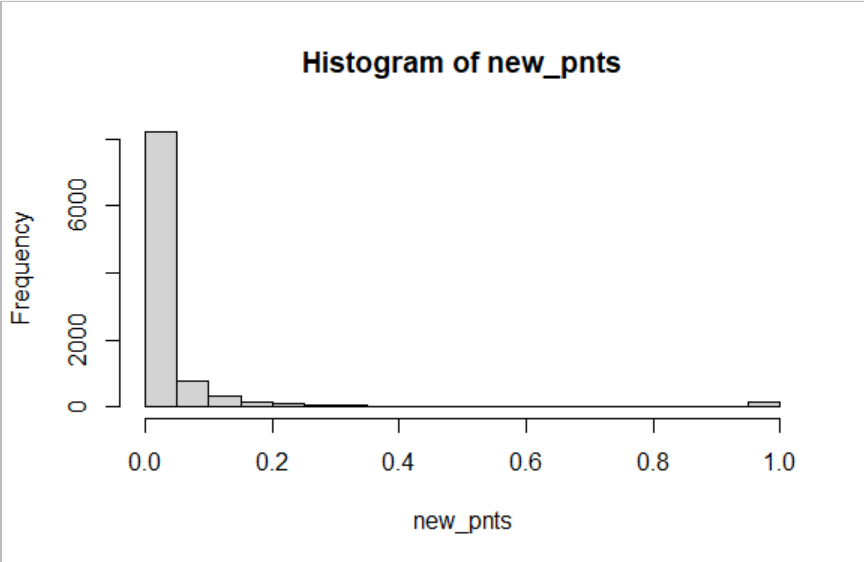
pnts <- runif(10000)
new_pnts <- sapply(pnts, flc_Inv)
Now you can use the function below to sample:
r_flc <- function(n) sapply(runif(n), flc_Inv)
hist(r_flc(10000))