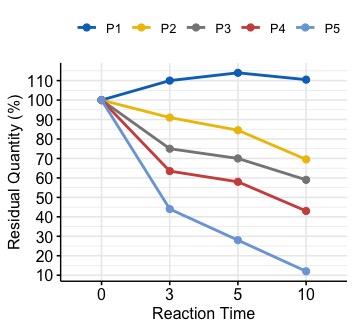
I have the following data frame:
df <- structure(list(
peptide = structure(c(
1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L
), levels = c(
"P1",
"P2", "P3", "P4", "P5"
), class = "factor"), reaction_time = c(
0,
3, 5, 10, 0, 3, 5, 10, 0, 3, 5, 10, 0, 3, 5, 10, 0, 3, 5, 10
),
mean_residual_quantity = c(
100, 110, 114, 110.5, 100, 91,
84.5, 69.5, 100, 75, 70, 59, 100, 63.5, 58, 43, 100, 44,
28, 12
)
), class = c("grouped_df", "tbl_df", "tbl", "data.frame"), row.names = c(NA, -20L), groups = structure(list(peptide = structure(1:5, levels = c(
"P1",
"P2", "P3", "P4", "P5"
), class = "factor"), .rows = structure(list(
1:4, 5:8, 9:12, 13:16, 17:20
), ptype = integer(0), class = c(
"vctrs_list_of",
"vctrs_vctr", "list"
))), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA, -5L), .drop = TRUE))
With this code:
ggpubr::ggline(df,
x = "reaction_time",
y = "mean_residual_quantity",
color = "peptide",
xlab = "Reaction Time",
palette = "jco",
size = 1,
ylab = "Residual Quantity (%)"
)
scale_y_continuous(breaks = get_breaks(n = 10))
grids()
rremove("legend.title")
I can create this plot:
Visually, we can see that the slope of the graph keeps decreasing from P1 to P5. Is there a single measure I can use to quantify this decreasing trend, for each P1 to P5?
So in the end, if we rank that value, the order should be the P1, P2, P3, P4, P5.
And how can I implement it with R?
CodePudding user response:
Suppose that slope is slope between first and last values.
If first mean_residual_quantity value of each peptide are same as your data,
library(dplyr)
df %>%
arrange(reaction_time) %>%
group_by(peptide) %>%
filter(row_number() == n()) %>%
arrange(desc(mean_residual_quantity)) %>% pull(peptide)
if not,
library(dplyr)
library(data.table)
df %>%
arrange(reaction_time) %>%
group_by(peptide) %>%
filter(row_number() == 1 | row_number() == n()) %>%
summarize(slope = (last(mean_residual_quantity) - first(mean_residual_quantity))/
(last(reaction_time) - first(reaction_time))) %>%
arrange(desc(slope)) %>% pull(peptide)
will give your desired result
[1] P1 P2 P3 P4 P5
Levels: P1 P2 P3 P4 P5
Here's another option that order by mean of slope between each two consecutive points.
df %>%
arrange(peptide, mean_residual_quantity) %>%
group_by(peptide) %>%
mutate(slope = (mean_residual_quantity - lag(mean_residual_quantity))/
(reaction_time - lag(reaction_time))) %>%
summarize(slope_mean = mean(slope, na.rm = T)) %>%
arrange(desc(slope_mean)) %>% pull(peptide)
CodePudding user response:
Another possible and completely arbitrary approach, rank each P for each time point, and then sum the ranks to obtain a sort of total rank. Transcribed your data from the graph
df=data.frame(
"RT"=c(0,3,5,10),
"P1"=c(100,110,115,110),
"P2"=c(100,90,85,70),
"P3"=c(100,75,70,60),
"P4"=c(100,65,60,40),
"P5"=c(100,45,30,10)
)
tmp=t(apply(df[,-1],1,rank))
colSums(tmp)
P1 P2 P3 P4 P5
18 15 12 9 6