| Pathway | #Proteins | Pvalue | Richfactor |
|---|---|---|---|
| Peptide chain elongation | 90 | 1.11E-16 | 0.5 |
| Translation elongation | 79 | 1.11E-16 | 0.7 |
| P53 pathway | 50 | 1.11E-16 | 0.2 |
| cGAS sting pathway | 20 | 1.11E-16 | 0.4 |
The above given is the data. Using this data i tried to generate bar graph with pvalue and proteins but i want to add additional details to graph like Rich factor given in the data above.
library(ggplot2)
library(viridis)
top_fun <- read.delim(file="Pathways.txt",header = TRUE)
topfun <- as.data.frame(top_fun)
#Turn your 'Name' column into a character vector
topfun$Pathway <- as.character(topfun$Pathway)
#Then turn it back into a factor with the levels in the correct order
topfun$Pathway<- factor(topfun$Pathway, levels=unique(topfun$Pathway))
ggplot(topfun,aes(x=Group,y=topfun$Proteins,fill=topfun$Pvalue))
geom_col(position="dodge",width=0.4)
coord_flip() scale_fill_viridis(option="mako")
facet_grid(Pathway~.)
theme(strip.text.y = element_text(angle = 0))
Using the above code i generated this graph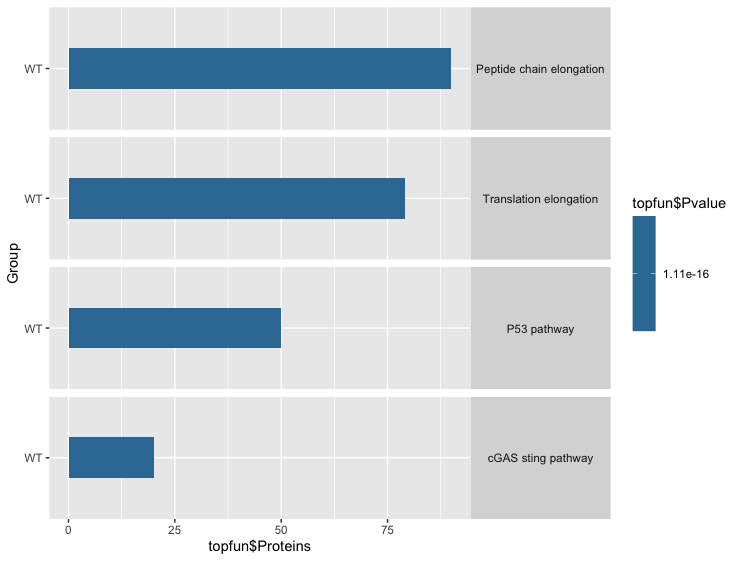
I want to add additional details like rich factor to the graph. Thanks for the help!.
CodePudding user response:
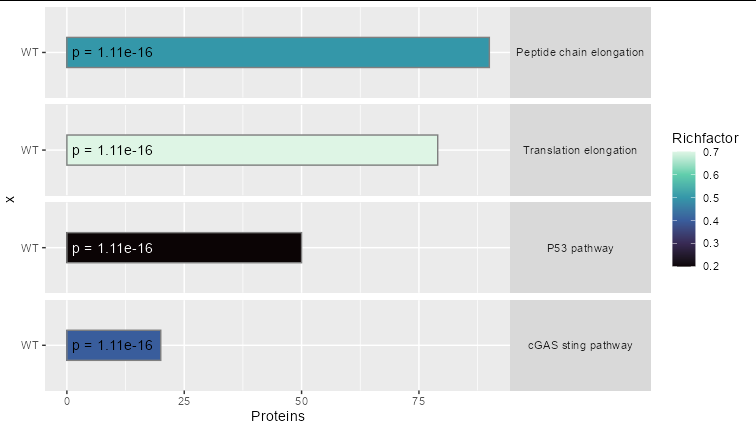
The obvious thing to do is to map Richfactor to the fill variable. You can add the p values directly as text, since they don't seem to be very helpful mapped to the fill scale, at least in this example
ggplot(topfun,aes(x = 'WT', y = Proteins, fill = Richfactor))
geom_col(position = "dodge", width = 0.4, color = 'gray50')
geom_text(aes(y = 1, label = paste('p =', Pvalue), color = Pathway),
hjust = 0)
coord_flip()
scale_fill_viridis_c(option = "mako")
facet_grid(Pathway ~ .)
theme(strip.text.y = element_text(angle = 0))
scale_color_manual(values = c('black', 'black', 'white', 'black'),
guide = 'none')