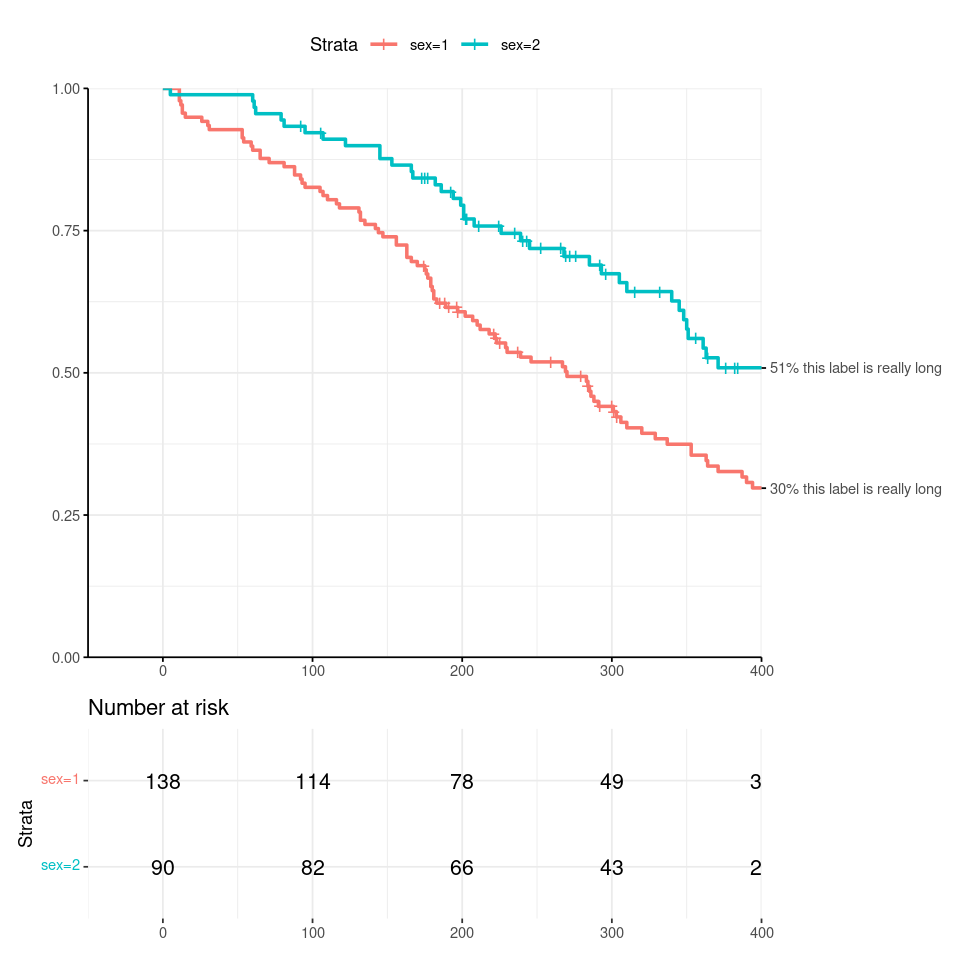
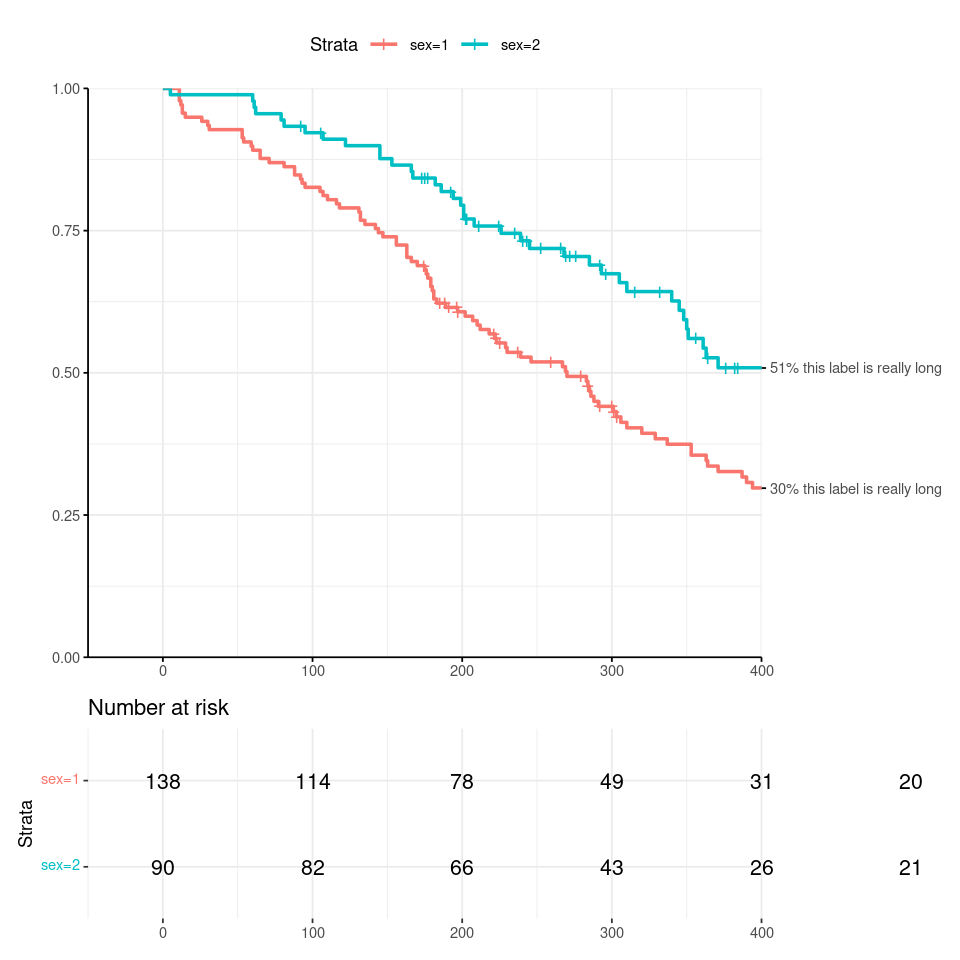
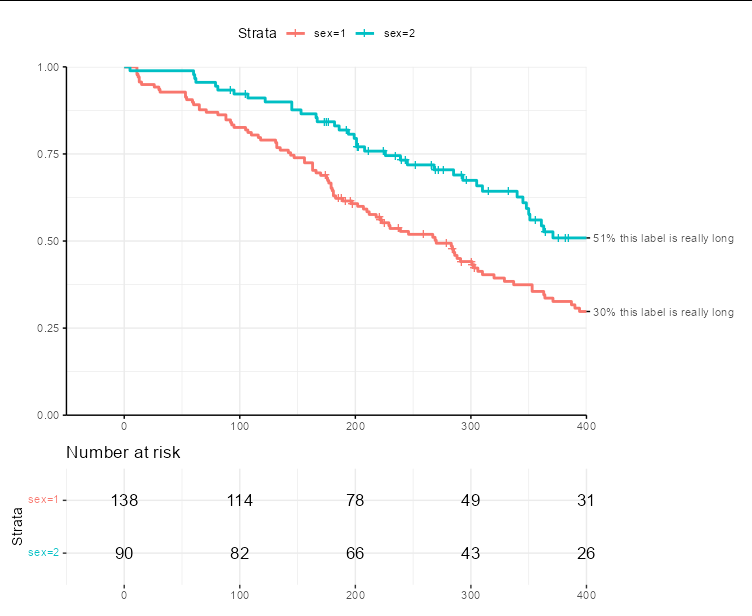
I want to specify the end time in my ggsurvplot, annotating the ends of the survival curves as shown below. But this leads to a truncation of the rightmost text in number-at-risk table. See code and output graphic below. Note that in the graphic, the rightmost values are 20 and 21, but all you see is 2 and 2.
If I increase the right-side x-axis limit (replace xlim=c(-50, time_cutoff) with xlim=c(-50, time_cutoff 50)), the risk table text is not clipped, but the survival curves are also plotted out to time_cutoff 50 and the labels I show to the right of the survival curve do not align with the ends of the survival curves.\
If I set clip='off' for just the risk table, I get extra values printed past the xlim cutoff.
library(survival)
library(survminer)
fit<- survfit(Surv(time, status) ~ sex, data = lung)
# Customized survival curves
my_plot = ggsurvplot(fit, data = lung,
# Add p-value and tervals
risk.table = TRUE,
tables.height = 0.2,
break.x.by=100,
ggtheme = theme_bw() # Change ggplot2 theme
)
time_cutoff = 400
xticks = c(0, 100, 200, 300, 400)
survs = summary(fit, times=time_cutoff)$surv
labels = paste(round(survs*100), '% this label is really long', sep='')
my_plot$plot <- my_plot$plot
coord_cartesian(ylim=c(0,1), xlim = c(-50,time_cutoff), clip = 'on', expand=FALSE)
scale_x_continuous(name=NULL, breaks = xticks)
scale_y_continuous(name=NULL, sec.axis=sec_axis(~., name=NULL, breaks = survs, labels= labels))
theme(
panel.border = element_blank(),
axis.line.y.left = element_line(color = 'black'),
axis.line.x.bottom = element_line(color = 'black'),
)
table_ylim = ggplot_build(my_plot$table)$layout$panel_params[[1]]$y.range
my_plot$table <- my_plot$table
coord_cartesian(ylim=table_ylim, xlim = c(-50,time_cutoff), clip = 'on', expand=FALSE)
scale_x_continuous(name=NULL, breaks = xticks)
theme(panel.border = element_blank())
library(patchwork)
(my_plot$plot / my_plot$table) plot_layout(heights = c(3,1))