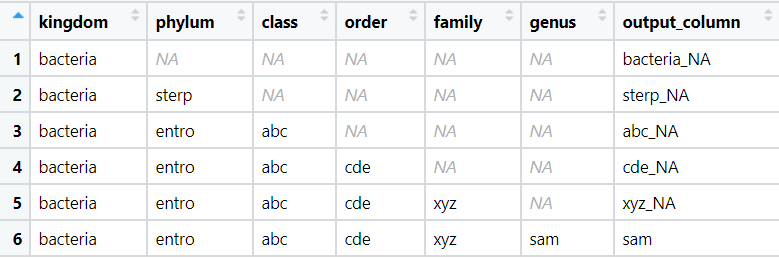
I am using R tidyverse and I have a tibble like the following code. I am trying to create the output_column based on the values of other columns. The data comes from the last non-empty column plus NA if there is an NA column before output_column.
library(tidyverse)
test_df <-
tibble(kingdom = rep("bacteria",6),
phylum = c(NA, "sterp", rep("entro", 4)),
class = c(rep(NA, 2), rep("abc",4)),
order= c(rep(NA,3), rep("cde", 3)),
family= c(rep(NA,4), rep("xyz", 2)),
genus= c(rep(NA,5), "sam"),
output_column = c("bacteria_NA", "sterp_NA", "abc_NA", "cde_NA", "xyz_NA", "sam" ))
CodePudding user response:
You can use row_wise() and c_across(), as follows:
test_df %>%
rowwise() %>%
mutate(k = if_else(!is.na(genus), genus, paste0(last(c_across()[!is.na(c_across())]), "_NA")))
Output:
kingdom phylum class order family genus k
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 bacteria NA NA NA NA NA bacteria_NA
2 bacteria sterp NA NA NA NA sterp_NA
3 bacteria entro abc NA NA NA abc_NA
4 bacteria entro abc cde NA NA cde_NA
5 bacteria entro abc cde xyz NA xyz_NA
6 bacteria entro abc cde xyz sam sam
Another approach is to use last() function with apply():
test_df$output_column = apply(
test_df, 1, \(x) {
if_else(is.na(last(x)), paste0(last(x[!is.na(x)]), "_NA"), last(x))
}
)
CodePudding user response:
We may use coalesce here
library(dplyr)
library(purrr)
library(stringr)
test_df %>%
mutate(output_column = invoke(coalesce, across(last_col():1)),
output_column = case_when(if_any((last_col()-1):1, is.na)~
str_c(output_column, '_NA'), TRUE ~ output_column))
-output
# A tibble: 6 × 7
kingdom phylum class order family genus output_column
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 bacteria <NA> <NA> <NA> <NA> <NA> bacteria_NA
2 bacteria sterp <NA> <NA> <NA> <NA> sterp_NA
3 bacteria entro abc <NA> <NA> <NA> abc_NA
4 bacteria entro abc cde <NA> <NA> cde_NA
5 bacteria entro abc cde xyz <NA> xyz_NA
6 bacteria entro abc cde xyz sam sam
data
test_df <- structure(list(kingdom = c("bacteria", "bacteria", "bacteria",
"bacteria", "bacteria", "bacteria"), phylum = c(NA, "sterp",
"entro", "entro", "entro", "entro"), class = c(NA, NA, "abc",
"abc", "abc", "abc"), order = c(NA, NA, NA, "cde", "cde", "cde"
), family = c(NA, NA, NA, NA, "xyz", "xyz"), genus = c(NA, NA,
NA, NA, NA, "sam")), row.names = c(NA, -6L), class = c("tbl_df",
"tbl", "data.frame"))