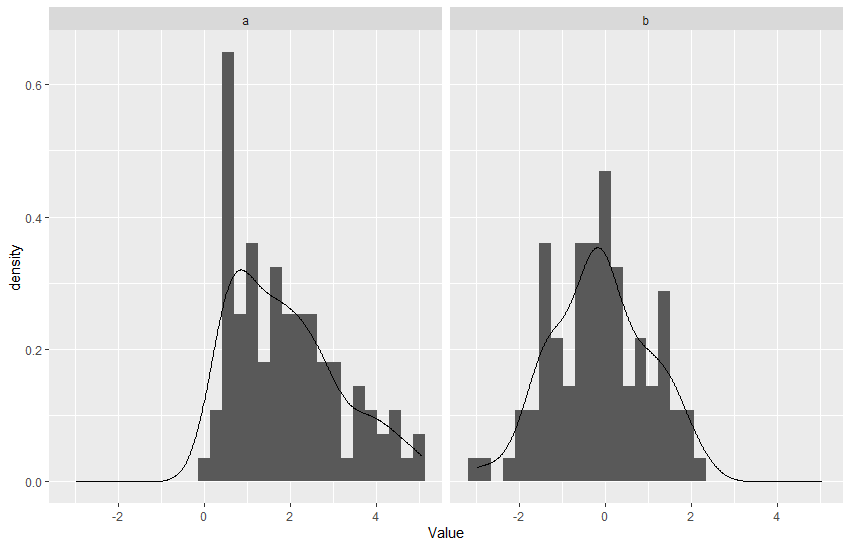
I'm using ggplot2 to create a faceted plot of the distributions of several variables for several groups, with a histogram and density estimate for each combination. Here is a simple, artificial example with one group and two variables:
library(tidyverse)
set.seed(42)
N <- 100
a <- rgamma(n = N, shape = 2)
b <- rnorm(n = N)
d <- tibble(x = 1:N, a = a, b = b) %>%
pivot_longer(cols = c("a", "b"), values_to = "Value", names_to = "Variable")
d %>% ggplot(mapping = aes(x = Value))
geom_histogram(mapping = aes(y = ..density..))
geom_density()
facet_wrap("Variable")
This gives
Note that a is gamma-distributed and therefore non-negative. (In my actual use case, although I of course don't know the distribution of the variables of interest, I know that some bounds exist for some of them.) I would like to take this into account for the kernel density estimate.
geom_density() has a bounds() argument that can be used for this, so if I were plotting a alone in the above example, I would use geom_density(bounds = c(0, Inf)). However, in the faceted plot, this would apply to the density estimates for both a and b, which is not what I want.
Is there a way (an easy one, ideally!) of setting per-variable bounds? I suspect that it may be possible and that I'll have to use stat_density() and geom_line() separately, but I'm unsure how to accomplish this. I have not found anything useful on Google or SO.
CodePudding user response:
This shows the general approach:
d %>% ggplot(mapping = aes(x = Value))
facet_wrap(~Variable)
geom_histogram(mapping = aes(y = ..density..))
geom_density(data = subset(d, Variable == "a"), trim = TRUE, colour = "red")
geom_density(data = subset(d, Variable == "b"), trim = FALSE)
Edit: For a beta distribution, look at using something like:
stat_function(data = subset(d, Variable == "a"),
fun = function(x) dbeta(**your parameters here**),
colour = "red",
size = 1)