I am trying to make a partial regression plot of a linear model, on which the y-axis shows the actual values vs x-axis but the displayed trend line is from the predict() function. As it is, my plot is showing jagged confidence intervals. How can I fix this? My model is looking at the effect of a metric of sleep disturbance on gene expression, controlling for age, sex, and education.
This is what I've done:
fit1<-lm(formula = (DF[, "gene.expression"]) ~ DF[, "sleep.metric"]
DF[, "age_death"] DF[, "msex"] DF[, "educ"])
pred<-fit1[["terms"]][[3]][[2]][[2]][[2]][[4]]
outcome<-fit1[["terms"]][[2]][[2]][[4]]
covars_list<-list(c('age_death', 'msex', 'educ'))
n<-length(fit1[["residuals"]])
b1slope<-coef(fit1)[2] #beta of sleep
x_var<-eval(parse(text=paste('DF$',pred,'[!is.na(DF$',pred,')]'))#values of sleep
b0intercept<-coef(fit1)['(Intercept)']
resid<-fit1$resid
y.fit<-fit1$fitted.values
#add back residuals to fitted vals to get actual y
y.adj<-b1slope*x b0intercept resid #gene expression
Since my data is scaled and centered, I make a new data frame containing values of sleep.metric and 0's for all other variables
covars_list<-list(c('age_death', 'msex', 'educ'))
terms<-c(pred, outcome, unlist(covars_list))
txt<-paste0('c("',paste(terms, collapse='","'),'")')
nf<-paste0('DF[,',txt,']', sep="")
nf<-eval(parse(text=nf))
nf[,2:dim(nf)[2]]=0
head(nf)
sleep.metric gene.exp age_death msex educ
45 0.07818673 0 0 0 0
81 0.13795502 0 0 0 0
131 -0.34721989 0 0 0 0
132 -0.74577821 0 0 0 0
189 -0.13113761 0 0 0 0
190 0.25137619 0 0 0 0
Then I predict the model on the new data frame:
p<-predict(fit1, newdata=data.frame(nf), se.fit=F, interval='confidence',level = 0.95)
p<-na.omit(p)
x<-x[order(x)]
y.adj<-y.adj[order(x)]
p<-p[order(x),]
fit<-p[,1]
lower<-p[,2]
upper<-p[,3]
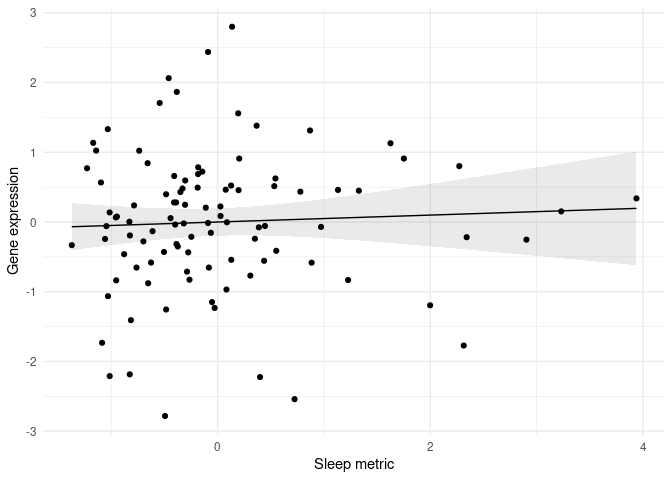
plot(y.adj~x,
pch=16, col='gray', cex=0.5,
xlab="A metric of sleep distrubances \n adjusted for Gene expression, Age, Sex, and Education",
ylab="Adjusted Normalized z-score of Differentially Expressed Genes")
lines(fit~x,col="blue", lty=1)
lines(upper~x, col="lightblue", lty=2)
lines(lower~x, col="lightblue", lty=2)
This is what the plot looks like. What is causing the error and how can I fix it?
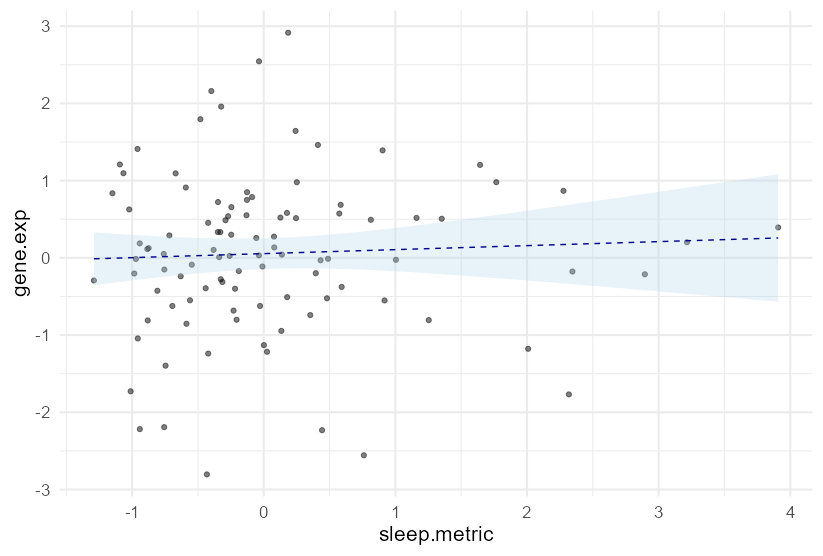
Note that you are effectively plotting the marginal effect of sleep.metric against the mean of the other covariates. In your case this results in a very similar plot to a straightforward geom_smooth(method = lm) of the two variables shown in your plot, so without any of the above code, you could just do:
ggplot(DF, aes(sleep.metric, gene.exp))
geom_point(alpha = 0.5)
geom_smooth(method = "lm", formula = y ~ x, fill = "lightblue",
color = "darkblue", linetype = 2, alpha = 0.3, linewidth = 0.5)
theme_minimal(base_size = 16)
CodePudding user response:
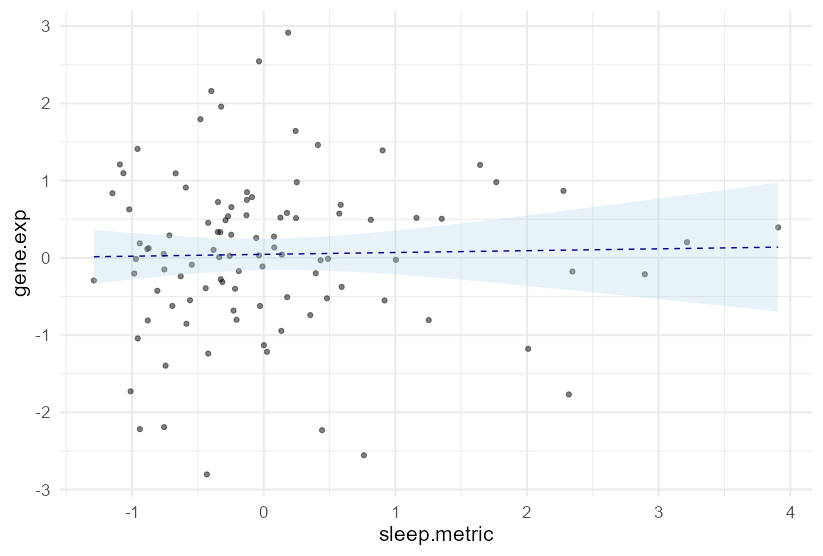
You could use the plot_cap() function from the marginaleffects package to do much of the heavy lifting for you. (Disclaimer: I am the maintainer.)
library(ggplot2)
library(marginaleffects)
# center
DF[] <- lapply(DF, scale)
DF[] <- lapply(DF, c)
# fit
fit1 <- lm(gene.exp ~ sleep.metric age_death msex educ, data = DF)
# plot
plot_cap(fit1, condition = "sleep.metric")
geom_point(data = DF, aes(sleep.metric, gene.exp))
labs(x = "Sleep metric", y = "Gene expression")
theme_minimal()