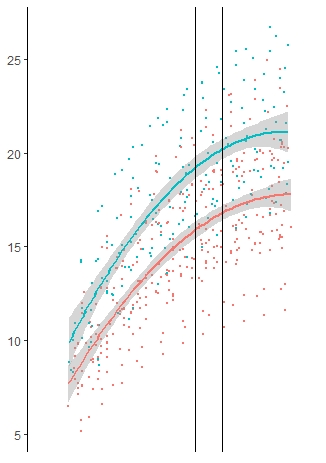
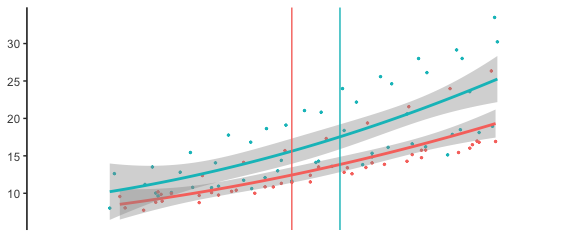
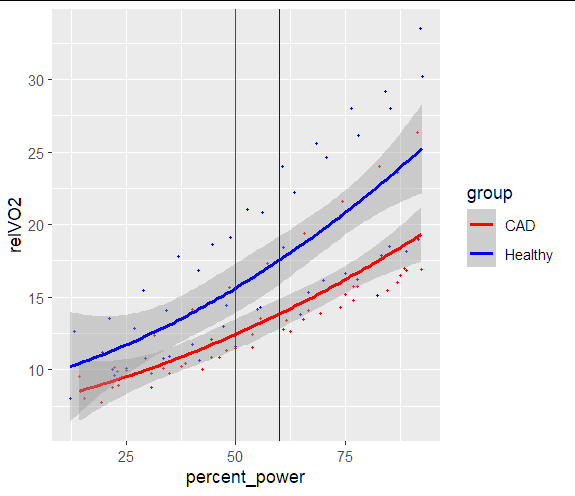
I have got this data:
structure(list(id = c("AC12-PRD-C1", "AC12-PRD-C1", "AC12-PRD-C1",
"AC12-PRD-C1", "AC12-PRD-C1", "AC12-PRD-C1", "AC12-PRD-C1", "AL13-PRD-C1",
"AL13-PRD-C1", "AL13-PRD-C1", "AL13-PRD-C1", "AL13-PRD-C1", "AL13-PRD-C1",
"AL13-PRD-C1", "AL13-PRD-C1", "BM06-PRD-S1", "BM06-PRD-S1", "BM06-PRD-S1",
"BM06-PRD-S1", "BM06-PRD-S1", "BM06-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1",
"CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1",
"CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1",
"CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1",
"CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1", "DA03-PRD-C1", "DA03-PRD-C1",
"DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1",
"DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1",
"DA24-PRD-S1", "DA24-PRD-S1", "DA24-PRD-S1", "DA24-PRD-S1", "DA24-PRD-S1",
"DA24-PRD-S1", "DA24-PRD-S1", "DB22-PRD-S1", "DB22-PRD-S1", "DB22-PRD-S1",
"DB22-PRD-S1", "DB22-PRD-S1", "DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1",
"DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1",
"DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1", "DL18-PRD-S1", "DL18-PRD-S1",
"DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1",
"DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1",
"DL18-PRD-S1", "DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1",
"DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1", "DT08-PRD-S1",
"DT08-PRD-S1", "DT08-PRD-S1", "DT08-PRD-S1", "DT08-PRD-S1", "DT08-PRD-S1",
"DT08-PRD-S1", "DT08-PRD-S1", "FB44-PRD-C1", "FB44-PRD-C1", "FB44-PRD-C1",
"FB44-PRD-C1", "FB44-PRD-C1", "FB44-PRD-C1", "FB44-PRD-C1"),
percent_power = c(21.7391304347826, 34.7826086956522, 47.8260869565217,
60.8695652173913, 73.9130434782609, 86.9565217391304, 100,
19.2307692307692, 30.7692307692308, 42.3076923076923, 53.8461538461538,
65.3846153846154, 76.9230769230769, 88.4615384615385, 100,
25, 40, 55, 70, 85, 100, 13.1578947368421, 21.0526315789474,
28.9473684210526, 36.8421052631579, 44.7368421052632, 52.6315789473684,
60.5263157894737, 68.4210526315789, 76.3157894736842, 84.2105263157895,
92.1052631578947, 100, 22.2222222222222, 33.3333333333333,
44.4444444444444, 55.5555555555556, 66.6666666666667, 77.7777777777778,
88.8888888888889, 100, 15.3846153846154, 23.0769230769231,
30.7692307692308, 38.4615384615385, 46.1538461538462, 53.8461538461538,
61.5384615384615, 69.2307692307692, 76.9230769230769, 84.6153846153846,
92.3076923076923, 100, 21.7391304347826, 34.7826086956522,
47.8260869565217, 60.8695652173913, 73.9130434782609, 86.9565217391304,
100, 29.4117647058824, 47.0588235294118, 64.7058823529412,
82.3529411764706, 100, 14.2857142857143, 22.8571428571429,
31.4285714285714, 40, 48.5714285714286, 57.1428571428571,
65.7142857142857, 74.2857142857143, 82.8571428571429, 91.4285714285714,
100, 12.1951219512195, 19.5121951219512, 26.8292682926829,
34.1463414634146, 41.4634146341463, 48.780487804878, 56.0975609756098,
63.4146341463415, 70.7317073170732, 78.0487804878049, 85.3658536585366,
92.6829268292683, 100, 22.2222222222222, 33.3333333333333,
44.4444444444444, 55.5555555555556, 66.6666666666667, 77.7777777777778,
88.8888888888889, 100, 41.6666666666667, 50, 58.3333333333333,
66.6666666666667, 75, 83.3333333333333, 91.6666666666667,
100, 25, 37.5, 50, 62.5, 75, 87.5, 100), relVO2 = c(8.79761904761905,
9.75892857142857, 11.3244047619048, 12.8005952380952, 14.2738095238095,
16.0208333333333, 17.8303571428571, 7.73391812865497, 8.75438596491228,
10, 11.5146198830409, 13.4444444444444, 15.7485380116959,
16.9707602339181, 18.6959064327485, 10.0941080196399, 11.7348608837971,
14.120294599018, 16.133387888707, 18.494271685761, 20.6546644844517,
12.6214285714286, 13.5107142857143, 15.4464285714286, 17.7714285714286,
18.6392857142857, 21.05, 23.9857142857143, 25.5821428571429,
28, 29.15, 33.4892857142857, 34.9071428571429, 9.63414634146342,
10.7479674796748, 12.0894308943089, 14.2886178861789, 15.3414634146341,
16.2073170731707, 18.1178861788618, 19.5569105691057, 8.04782608695652,
8.95, 9.71086956521739, 10.4108695652174, 10.85, 12.4021739130435,
13.4108695652174, 13.8630434782609, 14.754347826087, 15.4434782608696,
16.9152173913043, 17.7347826086956, 10.0187265917603, 10.9456928838951,
14.3960674157303, 18.3895131086142, 20.5945692883895, 23.5767790262172,
24.8408239700375, 10.7859281437126, 12.997754491018, 13.8248502994012,
15.1309880239521, 17.5561377245509, 9.56885456885457, 9.85521235521236,
12.3648648648649, 14.1248391248391, 15.6981981981982, 17.3133848133848,
19.3854568854569, 21.5637065637066, 23.9800514800515, 26.3384813384813,
27.5353925353925, 8.01307189542484, 11.1797385620915, 12.8104575163399,
14.0653594771242, 16.8235294117647, 19.1045751633987, 20.8267973856209,
22.1764705882353, 24.6274509803922, 26.1176470588235, 28.0065359477124,
30.2320261437909, 32.3398692810458, 10.1853015075377, 10.0942211055276,
10.8605527638191, 13.5175879396985, 14.0577889447236, 15.7663316582915,
16.8121859296482, 18.8316582914573, 10.6225425950197, 11.6186107470511,
13.5943643512451, 14.8623853211009, 16.6022280471822, 17.8604193971166,
18.9449541284404, 20.0360419397117, 9.98070987654321, 10.2546296296296,
11.4506172839506, 12.6041666666667, 15.1697530864198, 16.4930555555556,
17.8240740740741), group = c("CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD")), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -112L), groups = structure(list(
id = c("AC12-PRD-C1", "AL13-PRD-C1", "BM06-PRD-S1", "CB19-PRD-S1",
"CC14-PRD-S1", "DA03-PRD-C1", "DA24-PRD-S1", "DB22-PRD-S1",
"DB42-PRD-C1", "DL18-PRD-S1", "DR15-PRD-C1", "DT08-PRD-S1",
"FB44-PRD-C1"), .rows = structure(list(1:7, 8:15, 16:21,
22:33, 34:41, 42:53, 54:60, 61:65, 66:76, 77:89, 90:97,
98:105, 106:112), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -13L), .drop = TRUE))
I would like to get my vertical lines (geom_vline) the same color than the two graphs (50 as red for CAD, and 60 as blue for the healthy). This following code keeps my verticale lines black (see image)
Thank you!
varia_plot <- df_sum %>%
filter(percent_power < 100) %>%
ggplot(aes(x = percent_power, y = relVO2, color = group))
geom_point(size = 0.5)
geom_smooth(method = "lm", formula = y ~ poly(x, 2))
geom_vline(aes(color = group), xintercept = c(50, 60), linetype="solid")
xlab("Percentage of PPO (%)")
ylab(expression(paste("V", O[2]," (L/min)")))
scale_x_continuous(limits = c(0, 100), breaks = seq(0, 100, by = 25))
theme_classic()
theme(
panel.background = element_rect(fill ="white", colour = "white"),
axis.line = element_line(colour = "black"),
legend.title = element_text(face = "bold"),
legend.position = "none",
axis.line.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
strip.text = element_blank()
)
CodePudding user response:
Since you aren't using the main data.frame to draw the vline, it would be better to use a custom data= parameter on that layer. So use
geom_vline(aes(xintercept=val, color = group), linetype="solid",
data=data.frame(group=c("CAD", "Healthy"), val=c(50, 60)))
This will make things consistent with the global color= scale (as long as the values match exactly)
CodePudding user response:
What if you do like this: adding the color values to geom_vline:
geom_vline(color = c("red", "blue"), xintercept = c(50, 60), linetype="solid")
df %>%
filter(percent_power < 100) %>%
ggplot(aes(x = percent_power, y = relVO2, color = group))
geom_point(size = 0.5)
geom_smooth(method = "lm", formula = y ~ poly(x, 2))
geom_vline(color = c("red", "blue"), xintercept = c(50, 60), linetype="solid")
scale_color_manual(values = c("red", "blue"))
xlab("Percentage of PPO (%)")
ylab(expression(paste("V", O[2]," (L/min)")))
scale_x_continuous(limits = c(0, 100), breaks = seq(0, 100, by = 25))
theme_classic()
theme(
panel.background = element_rect(fill ="white", colour = "white"),
axis.line = element_line(colour = "black"),
legend.title = element_text(face = "bold"),
legend.position = "none",
axis.line.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
strip.text = element_blank()
)