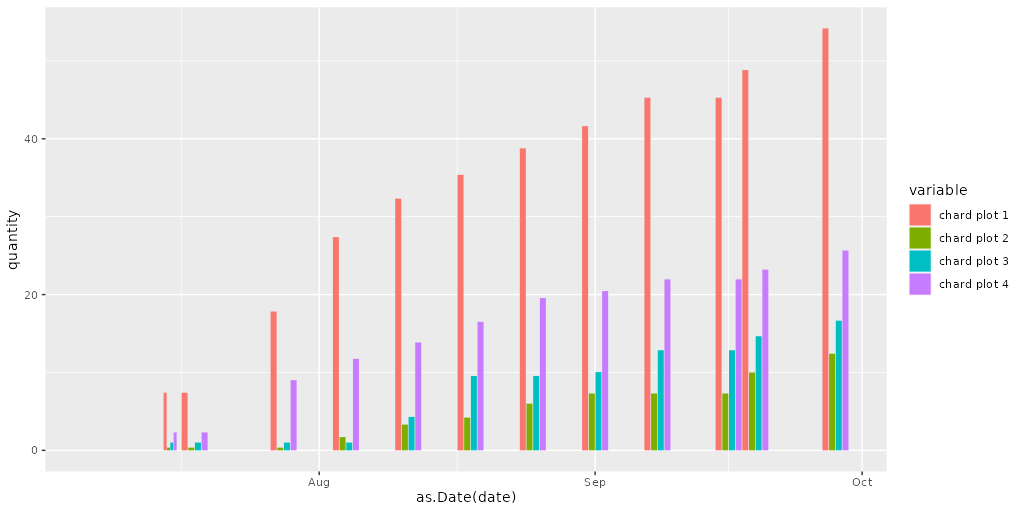
I get a position_dodge() requires non-overlapping x intervals` warning with the following data and code. If I omit the width = 3 option then I don't get the warning but the columns are almost impossible to distinguish. The plot of the ggplot code has lots of space between each set of columns so I wonder if anyone has a suggestion on what I need to change to get ride of the warning.
library(ggplot2)
g_crop <- ggplot(cumsum_2022_melt, aes(fill = variable, y = quantity, x = as.Date(date)))
geom_col(position="dodge", width = 3)
dput output
structure(list(date = structure(c(1657152000, 1657497600, 1657756800,
1657843200, 1658102400, 1658966400, 1659571200, 1660176000, 1660780800,
1661385600, 1661990400, 1662595200, 1663286400, 1663545600, 1664323200,
1657152000, 1657497600, 1657756800, 1657843200, 1658102400, 1658966400,
1659571200, 1660176000, 1660780800, 1661385600, 1661990400, 1662595200,
1663286400, 1663545600, 1664323200, 1657152000, 1657497600, 1657756800,
1657843200, 1658102400, 1658966400, 1659571200, 1660176000, 1660780800,
1661385600, 1661990400, 1662595200, 1663286400, 1663545600, 1664323200,
1657152000, 1657497600, 1657756800, 1657843200, 1658102400, 1658966400,
1659571200, 1660176000, 1660780800, 1661385600, 1661990400, 1662595200,
1663286400, 1663545600, 1664323200), class = c("POSIXct", "POSIXt"
), tzone = "UTC"), variable = c("chard plot 1", "chard plot 1",
"chard plot 1", "chard plot 1", "chard plot 1", "chard plot 1",
"chard plot 1", "chard plot 1", "chard plot 1", "chard plot 1",
"chard plot 1", "chard plot 1", "chard plot 1", "chard plot 1",
"chard plot 1", "chard plot 2", "chard plot 2", "chard plot 2",
"chard plot 2", "chard plot 2", "chard plot 2", "chard plot 2",
"chard plot 2", "chard plot 2", "chard plot 2", "chard plot 2",
"chard plot 2", "chard plot 2", "chard plot 2", "chard plot 2",
"chard plot 3", "chard plot 3", "chard plot 3", "chard plot 3",
"chard plot 3", "chard plot 3", "chard plot 3", "chard plot 3",
"chard plot 3", "chard plot 3", "chard plot 3", "chard plot 3",
"chard plot 3", "chard plot 3", "chard plot 3", "chard plot 4",
"chard plot 4", "chard plot 4", "chard plot 4", "chard plot 4",
"chard plot 4", "chard plot 4", "chard plot 4", "chard plot 4",
"chard plot 4", "chard plot 4", "chard plot 4", "chard plot 4",
"chard plot 4", "chard plot 4"), quantity = c(0, 0, 0, 7.4, 7.4,
17.82, 27.37, 32.32, 35.37, 38.77, 41.62, 45.27, 45.27, 48.82,
54.17, 0, 0, 0, 0.35, 0.35, 0.35, 1.7, 3.3, 4.2, 6, 7.3, 7.3,
7.3, 10, 12.4, 0, 0, 0, 1, 1, 1, 1, 4.3, 9.55, 9.55, 10.05, 12.85,
12.85, 14.65, 16.65, 0, 0, 0, 2.3, 2.3, 9, 11.75, 13.85, 16.5,
19.55, 20.45, 21.95, 21.95, 23.2, 25.65)), row.names = c(NA,
-60L), class = c("data.table", "data.frame"), .internal.selfref = <pointer: 0x135841ce0>, index = structure(integer(0), "`__variable`" = integer(0)))
CodePudding user response:
I don't know what the difference is, however I always use dodge2 instead of dodge. It seems to works in your situation as well:
Something funny is happening with the bars on the left, they are smaller then the rest of the bars. Maybe due to the records with zero data? This link talks about it: 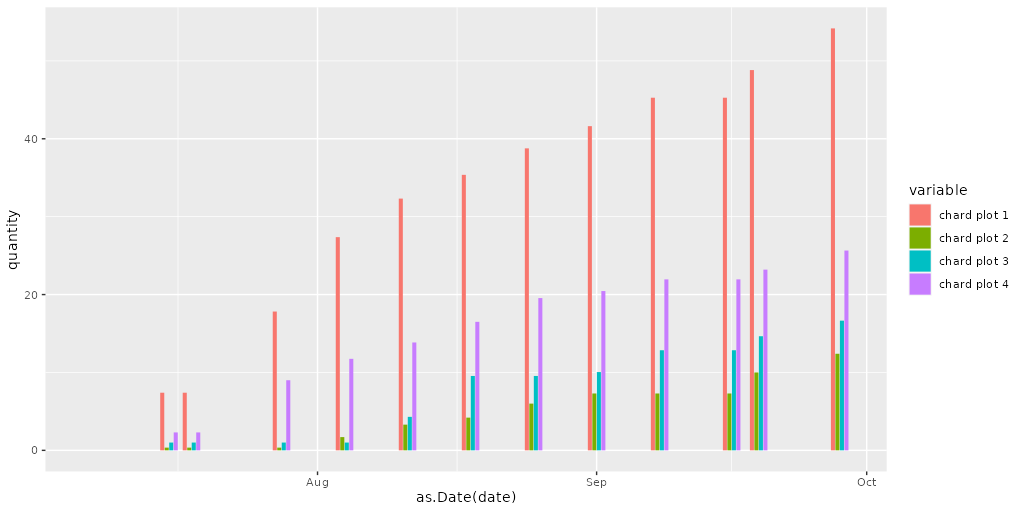
CodePudding user response:
The reason you are getting the warning is because ggplot2 observes that your data has daily resolution, in that it is not aligned to any longer time grid. As such, it will throw a warning when your dodging will cause a grouping to be wider than one day, since that risks the data having overlapping x intervals. It's a bit of false alarm here, since most of your groups are 7 days from their neighbors.
Since the visualization is otherwise what you want, the warning can be safely ignored.
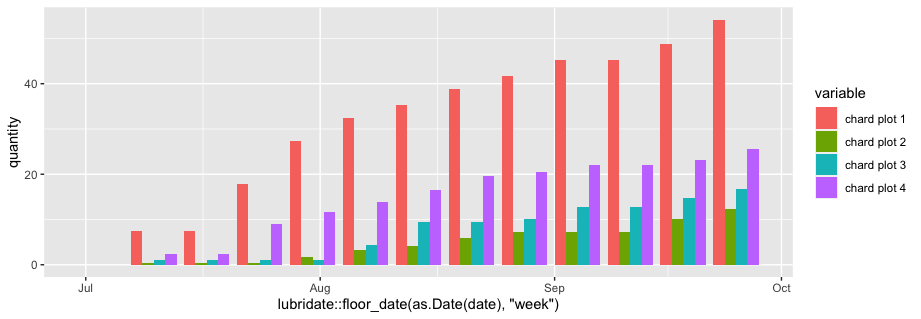
If you care about avoiding it, but you don't care about the precise alignment of your data to the specified dates, one option would be to align your data by week, such that ggplot recognizes that there are 7 days of width (ie width 7) that can be accommodated between groups. This doesn't throw a warning.
ggplot(cumsum_2022_melt, aes(fill = variable, y = quantity,
x = lubridate::floor_date(as.Date(date), "week")))
geom_col(position="dodge", width = 6)