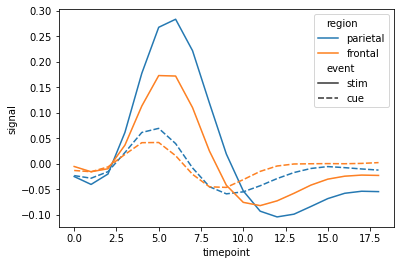
The plot in documentation looks like this :
 with code
with code
sns.lineplot(x="timepoint", y="signal",
hue="region", style="event",
data=fmri)
and
mine comes out to be like this
 for code :
for code :
sns.lineplot(
# data=fmri,
x=df["_C_UP"]["s"][:10],
y=df["_C_UP"]["px"][:10]
# hue="event"
);
How do I get the same effect for those lines ( that transparent color around it )
here is what my data looks like
#Energy s py pz px dxy dyz dz2 dxz dx2 tot
50 -17.98094 0.72320 0.31781 0.00000 0.31882 0.0 0.0 0.0 0.0 0.0 1.35982
51 -17.87394 0.29726 0.14415 0.00000 0.14491 0.0 0.0 0.0 0.0 0.0 0.58632
52 -17.76794 0.63694 0.02456 0.00000 0.02484 0.0 0.0 0.0 0.0 0.0 0.68634
53 -17.66194 1.78595 0.06032 0.00001 0.06139 0.0 0.0 0.0 0.0 0.0 1.90766
54 -17.55494 1.97809 0.09038 0.00001 0.09192 0.0 0.0 0.0 0.0 0.0 2.16040
CodePudding user response:
It depends on the data. The plot from the seaborn documentation that you show is based on a dataset where for every x value there are several y values (repeated measurements). The lines in the plot then indicate the means of those y values, and the shaded regions indicate the associated 95% confidence intervals.
In your data, there is only one y value for each x value, so there is no way to calculate a confidence interval.
CodePudding user response:
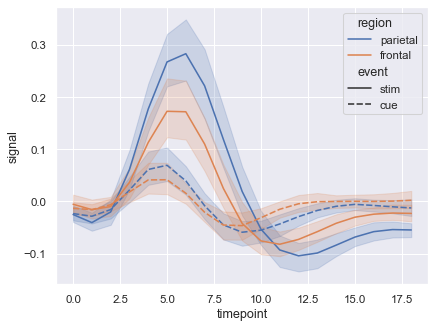
In the fmri datasets, there are actually multiple observations for each time point and subgroup, for example, at timepoint == 14 :
fmri[fmri['timepoint']==14]
subject timepoint event region signal
1 s5 14 stim parietal -0.080883
57 s13 14 stim parietal -0.033713
58 s12 14 stim parietal -0.068297
59 s11 14 stim parietal -0.114469
60 s10 14 stim parietal -0.052288
61 s9 14 stim parietal -0.130267
So the line you see, is actually the mean of all these observations (stratified by group) and the ribbon is the 95% confidence interval of this mean. For example, you can turn this off by doing:
sns.lineplot(x="timepoint", y="signal",
hue="region", style="event",
data=fmri,ci=None)
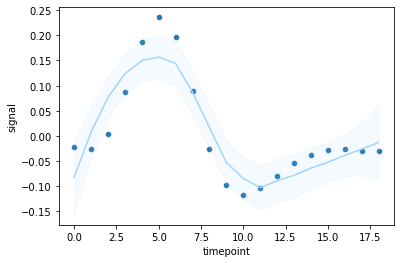
So to get the exact plot, you need to have multiple observations or replicates. If you don't, and your intention is to just connect the points, you cannot get a confidence interval.
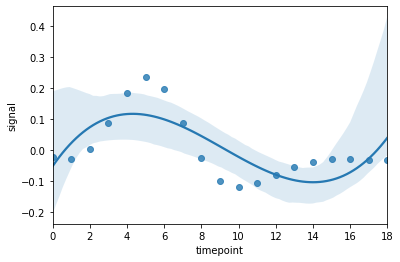
If you want to look at a trend line, one thing you can try is a polynomial smooth. And it makes sense to plot the data as points too.
Using an example from the same fmri dataset:
df = fmri[(fmri['subject']=="s5") & (fmri['event']== "stim") & (fmri['region'] == "frontal")]
sns.regplot(data=df,x = "timepoint",y = "signal",order=3)