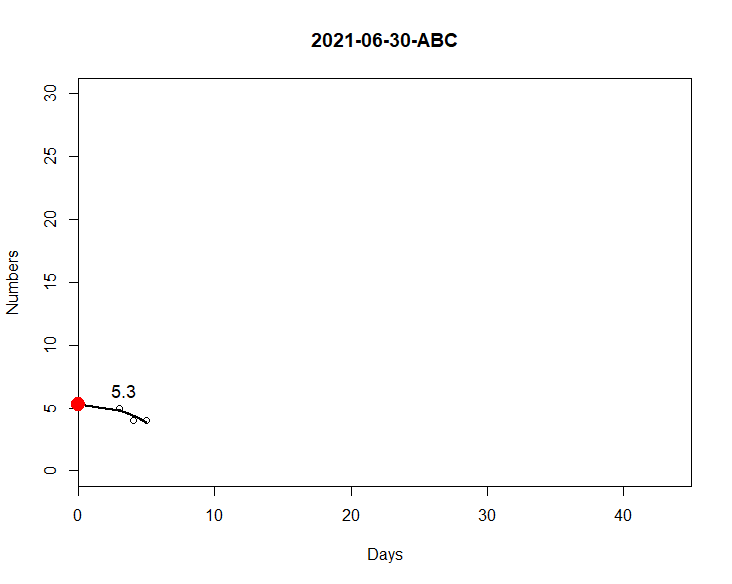
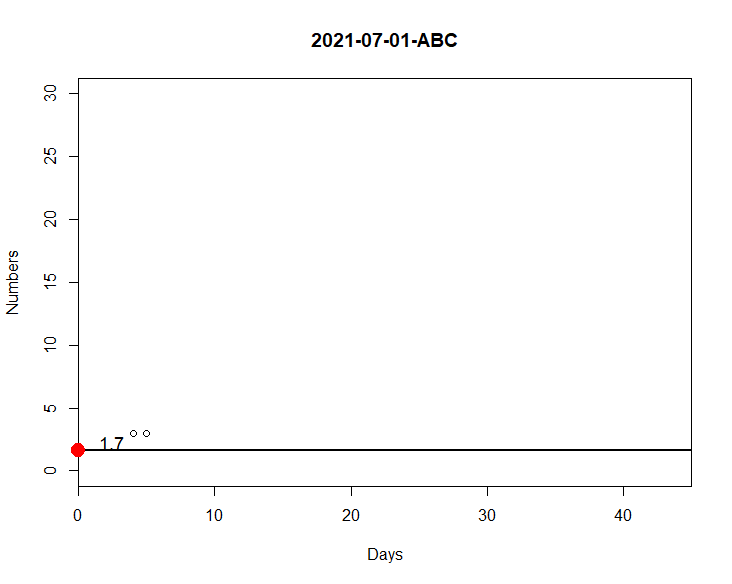
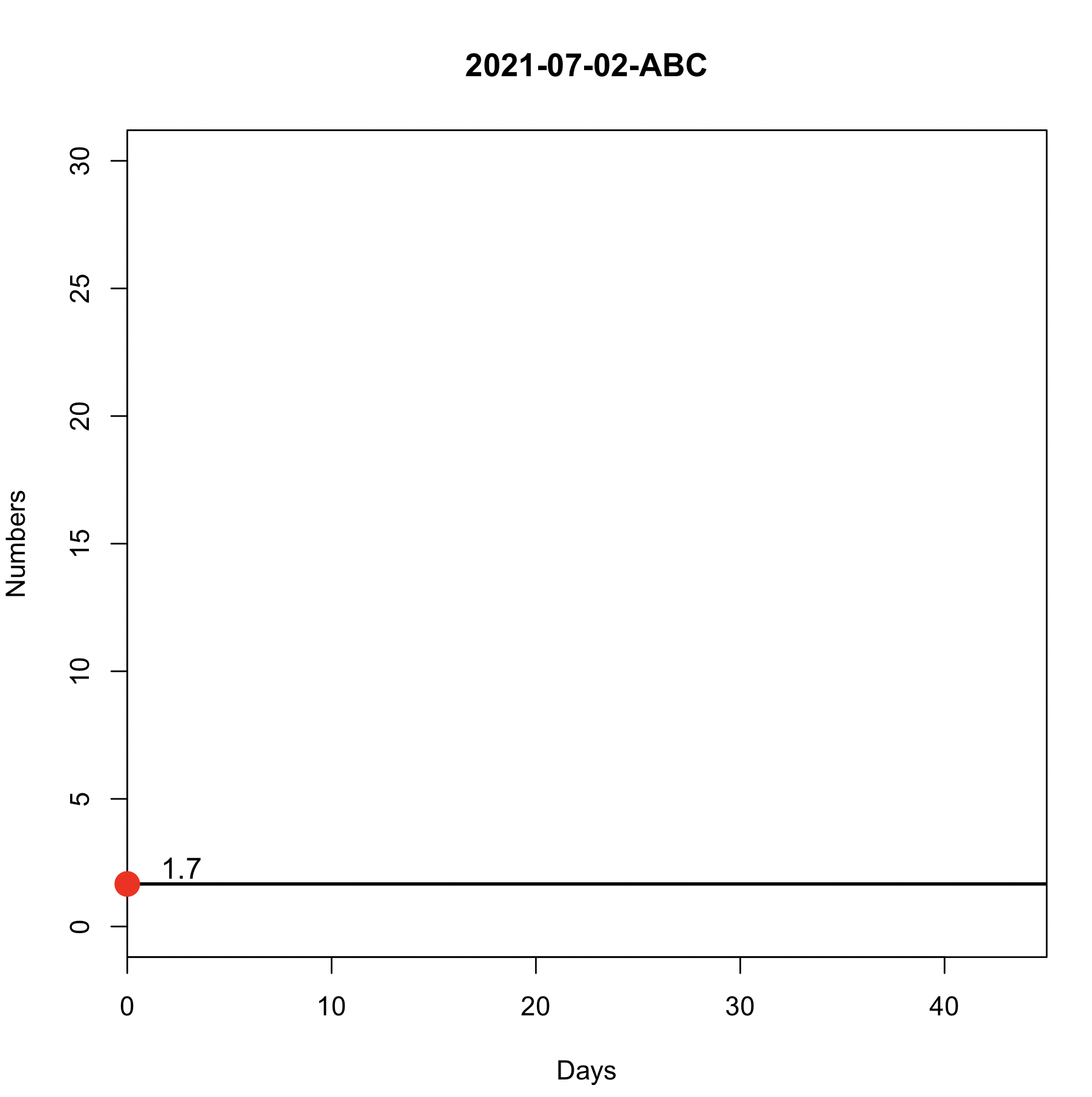
The code below generates a graph according to the day/category I choose on my date2. The days are 30/06, 01/07 and 02/07. For 30/06 and 01/07, I can generate normally as you can see in the attached image, but not for 02/07. This is because all my columns have 0 values and it ends up generating a problem in datas. So I need that if all columns are 0, I would like my graph to consider this condition of the code:
if (nrow(datas)<=2){
abline(h=m,lwd=2)
points(0, m, col = "red", pch = 19, cex = 2, xpd = TRUE)
text(.1,m .5, round(m,1), cex=1.1,pos=4,offset =1,col="black")}
So my graph would have no points, just the line in m.
Executable code below
library(dplyr)
df1 <- structure(
list(date1= c("2021-06-28","2021-06-28","2021-06-28"),
date2 = c("2021-06-30","2021-07-01","2021-07-02"),
Category = c("ABC","ABC","ABC"),
Week= c("Wednesday","Wednesday","Wednesday"),
DR1 = c(4,1,0),
DR01 = c(4,1,0), DR02= c(4,2,0),DR03= c(9,5,0),
DR04 = c(5,4,0),DR05 = c(5,4,0)),
class = "data.frame", row.names = c(NA, -3L))
f1 <- function(dmda, CategoryChosse) {
x<-df1 %>% select(starts_with("DR0"))
x<-cbind(df1, setNames(df1$DR1 - x, paste0(names(x), "_PV")))
PV<-select(x, date2,Week, Category, DR1, ends_with("PV"))
med<-PV %>%
group_by(Category,Week) %>%
summarize(across(ends_with("PV"), median))
SPV<-df1%>%
inner_join(med, by = c('Category', 'Week')) %>%
mutate(across(matches("^DR0\\d $"), ~.x
get(paste0(cur_column(), '_PV')),
.names = '{col}_{col}_PV')) %>%
select(date1:Category, DR01_DR01_PV:last_col())
SPV<-data.frame(SPV)
mat1 <- df1 %>%
filter(date2 == dmda, Category == CategoryChosse) %>%
select(starts_with("DR0")) %>%
pivot_longer(cols = everything()) %>%
arrange(desc(row_number())) %>%
mutate(cs = cumsum(value)) %>%
filter(cs == 0) %>%
pull(name)
(dropnames <- paste0(mat1,"_",mat1, "_PV"))
SPV <- SPV %>%
filter(date2 == dmda, Category == CategoryChosse) %>%
select(-any_of(dropnames))
datas<-SPV %>%
filter(date2 == ymd(dmda)) %>%
group_by(Category) %>%
summarize(across(starts_with("DR0"), sum)) %>%
pivot_longer(cols= -Category, names_pattern = "DR0(. )", values_to = "val") %>%
mutate(name = readr::parse_number(name))
colnames(datas)[-1]<-c("Days","Numbers")
datas <- datas %>%
group_by(Category) %>%
slice((as.Date(dmda) - min(as.Date(df1$date1) [
df1$Category == first(Category)])):max(Days) 1) %>%
ungroup
plot(Numbers ~ Days, xlim= c(0,45), ylim= c(0,30),
xaxs='i',data = datas,main = paste0(dmda, "-", CategoryChosse))
m<-df1 %>%
group_by(Category,Week) %>%
summarize(across(starts_with("DR1"), mean))
m<-subset(m, Week == df1$Week[match(ymd(dmda), ymd(df1$date2))] & Category == CategoryChosse)$DR1
if (nrow(datas)<=2){
abline(h=m,lwd=2)
points(0, m, col = "red", pch = 19, cex = 2, xpd = TRUE)
text(.1,m .5, round(m,1), cex=1.1,pos=4,offset =1,col="black")}
else if(any(table(datas$Numbers) >= 3) & length(unique(datas$Numbers)) == 1){
yz <- unique(datas$Numbers)
lines(c(0,datas$Days), c(yz, datas$Numbers), lwd = 2)
points(0, yz, col = "red", pch = 19, cex = 2, xpd = TRUE)
text(.1,yz .5,round(yz,1), cex=1.1,pos=4,offset =1,col="black")}
else{
mod <- nls(Numbers ~ b1*Days^2 b2,start = list(b1 = 0,b2 = 0),data = datas, algorithm = "port")
new.data <- data.frame(Days = with(datas, seq(min(Days),max(Days),len = 45)))
new.data <- rbind(0, new.data)
lines(new.data$Days,predict(mod,newdata = new.data),lwd=2)
coef<-coef(mod)[2]
points(0, coef, col="red",pch=19,cex = 2,xpd=TRUE)
text(.99,coef 1,max(0, round(coef,1)), cex=1.1,pos=4,offset =1,col="black")
}
}
f1("2021-06-30", "ABC")
f1("2021-07-01", "ABC")
f1("2021-07-02", "ABC")
CodePudding user response:
The DR0 columns are removed in the last case and this results in error because the summarise is looping through those columns summarize(across(starts_with("DR0"), sum)). An option is to create a condition check i.e. if there are no DR0 columns left then add those columns as NA and it should work without any error
f1 <- function(dmda, CategoryChosse) {
x<-df1 %>% select(starts_with("DR0"))
x<-cbind(df1, setNames(df1$DR1 - x, paste0(names(x), "_PV")))
PV<-select(x, date2,Week, Category, DR1, ends_with("PV"))
med<-PV %>%
group_by(Category,Week) %>%
summarize(across(ends_with("PV"), median))
SPV<-df1%>%
inner_join(med, by = c('Category', 'Week')) %>%
mutate(across(matches("^DR0\\d $"), ~.x
get(paste0(cur_column(), '_PV')),
.names = '{col}_{col}_PV')) %>%
select(date1:Category, DR01_DR01_PV:last_col())
SPV<-data.frame(SPV)
mat1 <- df1 %>%
filter(date2 == dmda, Category == CategoryChosse) %>%
select(starts_with("DR0")) %>%
pivot_longer(cols = everything()) %>%
arrange(desc(row_number())) %>%
mutate(cs = cumsum(value)) %>%
filter(cs == 0) %>%
pull(name)
(dropnames <- paste0(mat1,"_",mat1, "_PV"))
SPV <- SPV %>%
filter(date2 == dmda, Category == CategoryChosse) %>%
select(-any_of(dropnames))
if(length(grep("DR0", names(SPV))) == 0) {
SPV[mat1] <- NA_real_
}
datas <-SPV %>%
filter(date2 == ymd(dmda)) %>%
group_by(Category) %>%
summarize(across(starts_with("DR0"), sum)) %>%
pivot_longer(cols= -Category, names_pattern = "DR0(. )", values_to = "val") %>%
mutate(name = readr::parse_number(name))
colnames(datas)[-1]<-c("Days","Numbers")
datas <- datas %>%
group_by(Category) %>%
slice((as.Date(dmda) - min(as.Date(df1$date1) [
df1$Category == first(Category)])):max(Days) 1) %>%
ungroup
plot(Numbers ~ Days, xlim= c(0,45), ylim= c(0,30),
xaxs='i',data = datas,main = paste0(dmda, "-", CategoryChosse))
m<-df1 %>%
group_by(Category,Week) %>%
summarize(across(starts_with("DR1"), mean))
m<-subset(m, Week == df1$Week[match(ymd(dmda), ymd(df1$date2))] & Category == CategoryChosse)$DR1
if (nrow(datas)<=2){
abline(h=m,lwd=2)
points(0, m, col = "red", pch = 19, cex = 2, xpd = TRUE)
text(.1,m .5, round(m,1), cex=1.1,pos=4,offset =1,col="black")}
else if(any(table(datas$Numbers) >= 3) & length(unique(datas$Numbers)) == 1){
yz <- unique(datas$Numbers)
lines(c(0,datas$Days), c(yz, datas$Numbers), lwd = 2)
points(0, yz, col = "red", pch = 19, cex = 2, xpd = TRUE)
text(.1,yz .5,round(yz,1), cex=1.1,pos=4,offset =1,col="black")}
else{
mod <- nls(Numbers ~ b1*Days^2 b2,start = list(b1 = 0,b2 = 0),data = datas, algorithm = "port")
new.data <- data.frame(Days = with(datas, seq(min(Days),max(Days),len = 45)))
new.data <- rbind(0, new.data)
lines(new.data$Days,predict(mod,newdata = new.data),lwd=2)
coef<-coef(mod)[2]
points(0, coef, col="red",pch=19,cex = 2,xpd=TRUE)
text(.99,coef 1,max(0, round(coef,1)), cex=1.1,pos=4,offset =1,col="black")
}
}
-testing
f1("2021-07-02", "ABC")
-output