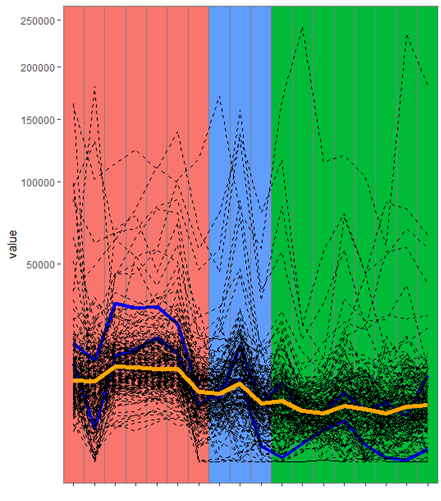
Hi My data (data_long) looks like this:
genes sample value Group Type
A1 O7high 6796.448 G0 A
AA O7high 4997.250 G0 A
A3 O7high 9477.100 G0 A
A4 O7high 6083.558 G0 A
A1 08low 075.364 G0 B
AA 08low 13066.130 G0 B
p <- ggplot(data_long, aes(x=sample, y=value,group=genes,color=Group))
geom_tile(aes(fill = as.factor(Type),color = NA,y = 7000), height = Inf, alpha = 0.5)
geom_line(aes(linetype=Group,color=Group, size=Group))
stat_summary(aes(group = -1), fun=median, geom='line',size=2, color='orange')
theme_classic()
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
scale_y_sqrt()
scale_colour_manual(values=c("black","blue"))
scale_size_manual(values=c(0.3,1.5))
scale_linetype_manual(values=c("dashed", "solid"))
theme_classic()
p theme_bw()
theme(panel.grid = element_blank(),
panel.border = element_blank())
I have used the above code to plot the median line as well as to highlight some of the genes of my interest. Everything works fine but When seeing the plot there are these vertical grey lines (Separating each sample?) I am not sure how to remove those lines. I want the geom_tile for the same type to be without any lines. Please let me know how to remove those lines
CodePudding user response:
Thanks for updating your question with the required info. Perhaps you can remove the grey lines by moving color = NA outside aes(), e.g.
library(tidyverse)
data_long <- read.table(text = "genes sample value Group Type
A1 O7med 6796.448 G0 A
AA O7med 4997.250 G0 A
A3 O7high 9477.100 G0 A
A4 O7high 6083.558 G0 A
A1 08low 075.364 G0 B
AA 08low 13066.130 G0 B", header = TRUE)
p <- ggplot(data_long, aes(x=sample, y=value,group=genes,color=Group))
geom_tile(aes(fill = as.factor(Type), y = 7000), color = NA, height = Inf, alpha = 0.5)
geom_line(aes(linetype=Group,color=Group, size=Group))
stat_summary(aes(group = -1), fun=median, geom='line',size=2, color='orange')
theme_classic()
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
scale_y_sqrt()
scale_colour_manual(values=c("black","blue"))
scale_size_manual(values=c(0.3,1.5))
scale_linetype_manual(values=c("dashed", "solid"))
p theme_bw()
theme(panel.grid = element_blank(),
panel.border = element_blank())
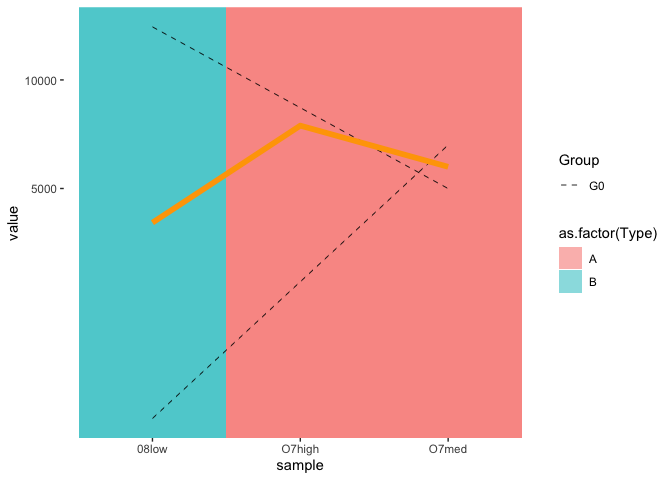
Created on 2021-10-13 by the reprex package (v2.0.1)
Does that work on your actual dataset?