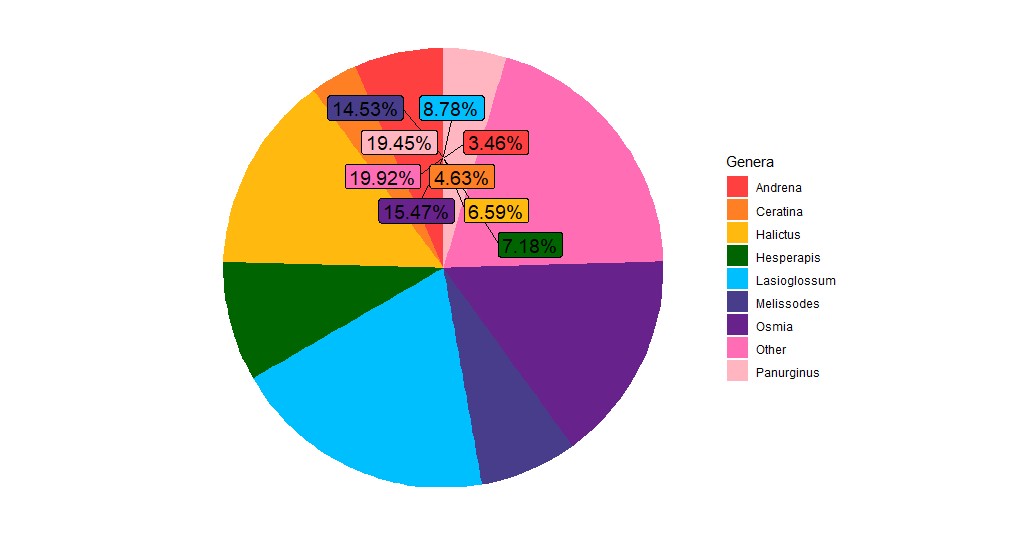
I am trying to create a pie chart to visualize percent abundance of 9 genera. However, the labels are all clumping together. How do I remedy this? Code included below:
generaabundance2020 <- c(883, 464, 1948, 1177, 2607, 962, 2073, 620, 2670)
genera2020 <- c("Andrena", "Ceratina", "Halictus",
"Hesperapis", "Lasioglossum", "Melissodes",
"Osmia", "Panurginus", "Other")
generabreakdown2020 <- data.frame(group = genera2020, value = generaabundance2020)
gb2020label <- generabreakdown2020 %>%
group_by(value) %>% # Variable to be transformed
count() %>%
ungroup() %>%
mutate(perc = `value` / sum(`value`)) %>%
arrange(perc) %>%
mutate(labels = scales::percent(perc))
generabreakdown2020 %>%
ggplot(aes(x = "", y = value, fill = group))
geom_col()
coord_polar("y", start = 0)
theme_void()
geom_label_repel(aes(label = gb2020label$labels), position = position_fill(vjust = 0.5),
size = 5, show.legend = F, max.overlaps = 50)
guides(fill = guide_legend(title = "Genera"))
scale_fill_manual(values = c("brown1", "chocolate1",
"darkgoldenrod1", "darkgreen",
"deepskyblue", "darkslateblue",
"darkorchid4", "hotpink1",
"lightpink"))
CodePudding user response:
You didn't provide us with your data to work with so I'm using ggplot2::mpg here.
library(tidyverse)
library(ggrepel)
mpg_2 <-
mpg %>%
slice_sample(n = 20) %>%
count(manufacturer) %>%
mutate(perc = n / sum(n)) %>%
mutate(labels = scales::percent(perc)) %>%
arrange(desc(manufacturer)) %>%
mutate(text_y = cumsum(n) - n/2)
Chart without polar coordinates
mpg_2 %>%
ggplot(aes(x = "", y = n, fill = manufacturer))
geom_col()
geom_label(aes(label = labels, y = text_y))
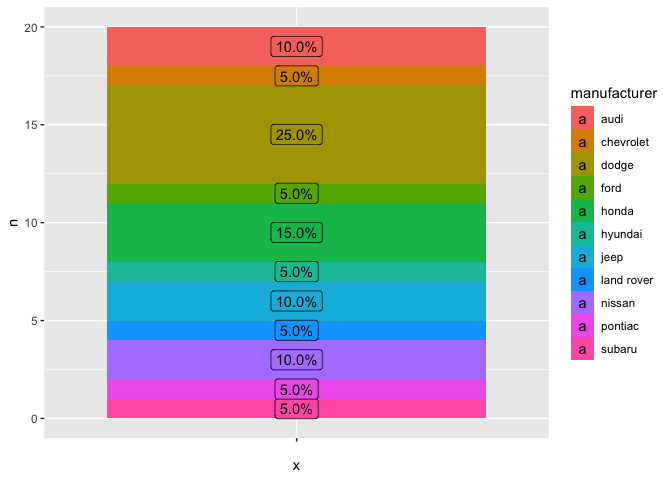
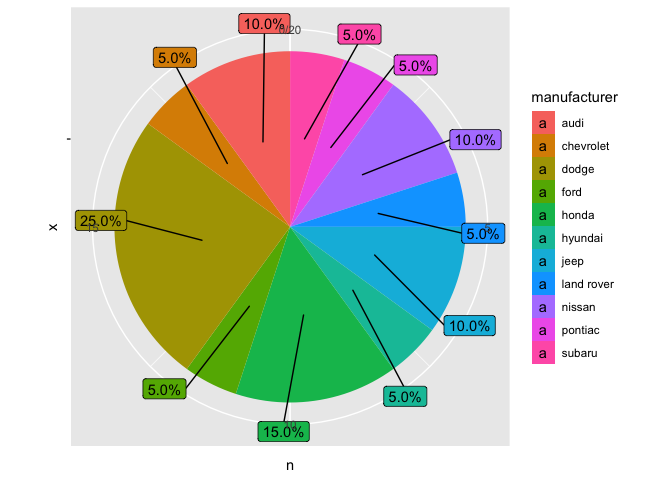
Chart with polar coordinates and geom_label_repel
mpg_2 %>%
ggplot(aes(x = "", y = n, fill = manufacturer))
geom_col()
geom_label_repel(aes(label = labels, y = text_y),
force = 0.5,nudge_x = 0.6, nudge_y = 0.6)
coord_polar(theta = "y")
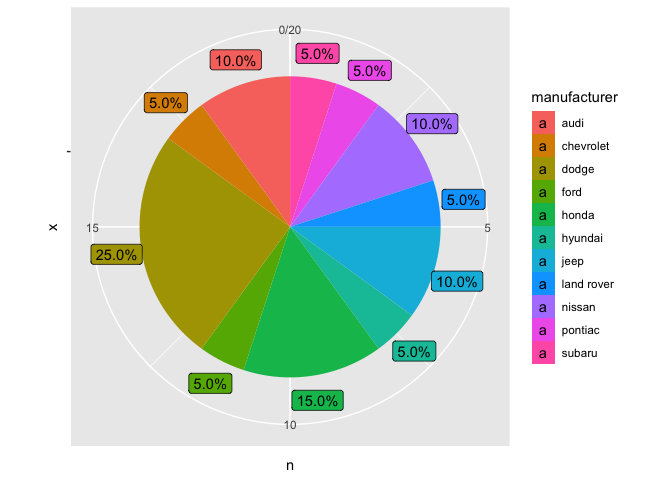
But maybe your data isn’t dense enough to need repelling?
mpg_2 %>%
ggplot(aes(x = "", y = n, fill = manufacturer))
geom_col()
geom_label(aes(label = labels, y = text_y), nudge_x = 0.6)
coord_polar(theta = "y")
Created on 2021-10-26 by the reprex package (v2.0.1)
CodePudding user response:
Thanks for adding your data.
There are a few errors in your code. The main one is that you didn't precalculate where to place the labels (done here in the text_y variable). That variable needs to be passed as the y aesthetic for geom_label_repel.
The second is that you no longer need
group_by(value) %>% count() %>% ungroup() because the data you provided is already aggregated.
library(tidyverse)
library(ggrepel)
generaabundance2020 <- c(883, 464, 1948, 1177, 2607, 962, 2073, 620, 2670)
genera2020 <- c("Andrena", "Ceratina", "Halictus", "Hesperapis", "Lasioglossum", "Melissodes", "Osmia", "Panurginus", "Other")
generabreakdown2020 <- data.frame(group = genera2020, value = generaabundance2020)
gb2020label <-
generabreakdown2020 %>%
mutate(perc = value/ sum(value)) %>%
mutate(labels = scales::percent(perc)) %>%
arrange(desc(group)) %>% ## arrange in the order of the legend
mutate(text_y = cumsum(value) - value/2) ### calculate where to place the text labels
gb2020label %>%
ggplot(aes(x = "", y = value, fill = group))
geom_col()
coord_polar(theta = "y")
geom_label_repel(aes(label = labels, y = text_y),
nudge_x = 0.6, nudge_y = 0.6,
size = 5, show.legend = F)
guides(fill = guide_legend(title = "Genera"))
scale_fill_manual(values = c("brown1", "chocolate1",
"darkgoldenrod1", "darkgreen",
"deepskyblue", "darkslateblue",
"darkorchid4", "hotpink1",
"lightpink"))
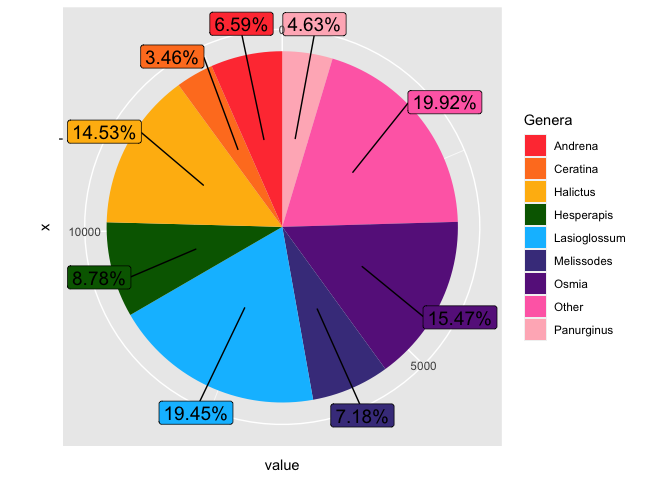
If you want to arrange in descending order of frequency, you should remember to also set the factor levels of the group variable to the same order.
gb2020label <-
generabreakdown2020 %>%
mutate(perc = value/ sum(value)) %>%
mutate(labels = scales::percent(perc)) %>%
arrange(desc(perc)) %>% ## arrange in descending order of frequency
mutate(group = fct_rev(fct_inorder(group))) %>% ## also arrange the groups in descending order of freq
mutate(text_y = cumsum(value) - value/2) ### calculate where to place the text labels
gb2020label %>%
ggplot(aes(x = "", y = value, fill = group))
geom_col()
coord_polar(theta = "y")
geom_label_repel(aes(label = labels, y = text_y),
nudge_x = 0.6, nudge_y = 0.6,
size = 5, show.legend = F)
guides(fill = guide_legend(title = "Genera"))
scale_fill_manual(values = c("brown1", "chocolate1",
"darkgoldenrod1", "darkgreen",
"deepskyblue", "darkslateblue",
"darkorchid4", "hotpink1",
"lightpink"))
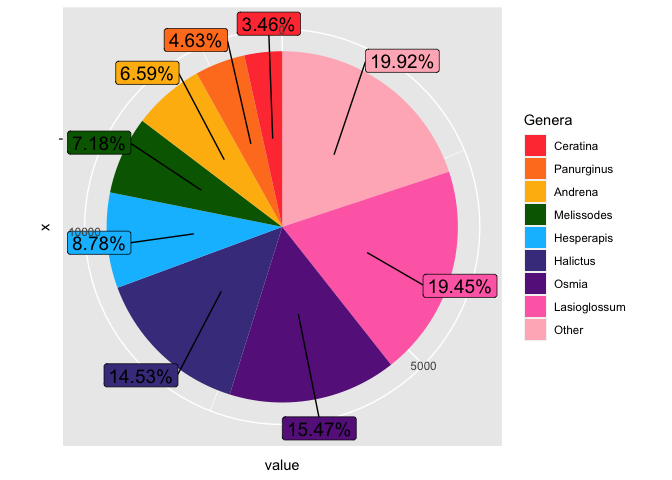
Created on 2021-10-27 by the reprex package (v2.0.1)