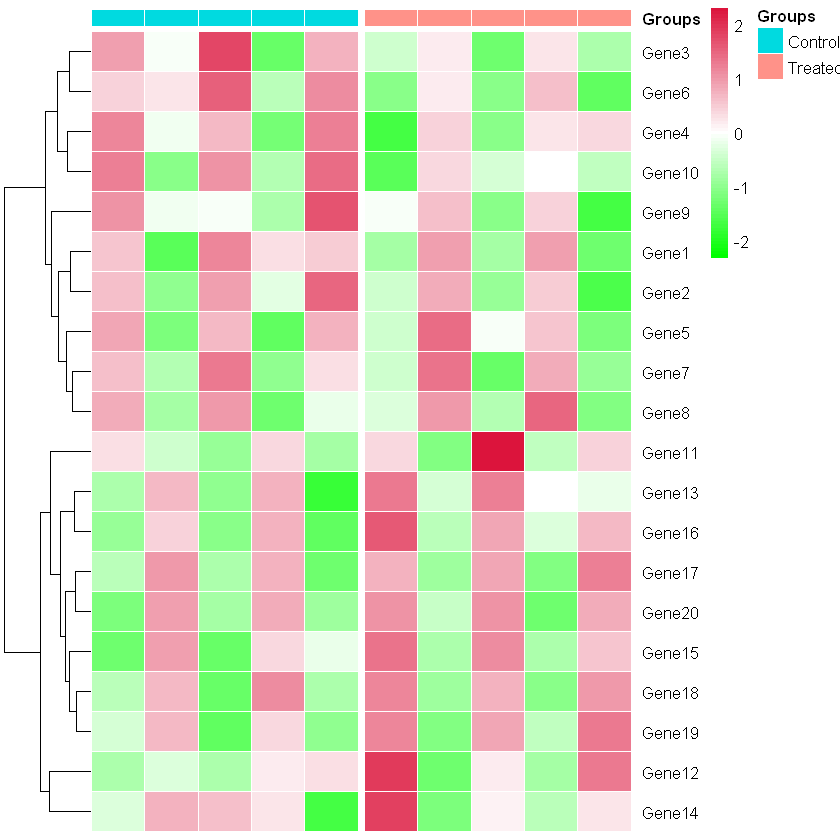
Consider the matrix:
test = matrix(rnorm(200), 20, 10)
test[1:10, seq(1, 10, 2)] = test[1:10, seq(1, 10, 2)] 3
test[11:20, seq(2, 10, 2)] = test[11:20, seq(2, 10, 2)] 2
test[15:20, seq(2, 10, 2)] = test[15:20, seq(2, 10, 2)] 4
colnames(test) = paste("Test", 1:10, sep = "")
rownames(test) = paste("Gene", 1:20, sep = "")
#creating group to divide
group_df = data.frame(Groups=rep(c("Control", "Treated"), c(5,5)))
rownames(group_df) <- colnames(test)
#Generating heatmap:
pheatmap(test, cluster_cols = FALSE, scale = 'row',
annotation_col = group_df,
show_colnames = FALSE,
border_color = "white",
colorRampPalette(c("#00FF00", "white", "#DC143C"))(75),
gaps_col = cumsum(c(5,5)))
How do I change the color of Control and Treated from blue and red to different colors?
CodePudding user response:
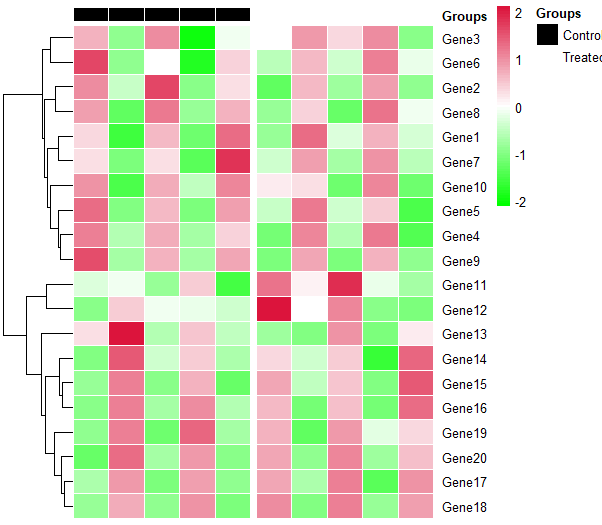
Change group_df to a factor and specifying annotation_colors will work.
group_df = data.frame(Groups=as.factor(rep(c("Control", "Treated"), c(5,5))))
ann_colors = list(
Groups = c(Control="black", Treated="white"))
rownames(group_df) <- colnames(dummy)
pheatmap(dummy, cluster_cols = FALSE, scale = 'row',
annotation_col = group_df,
annotation_colors = ann_colors,
show_colnames = FALSE,
border_color = "white",
colorRampPalette(c("#00FF00", "white", "#DC143C"))(75),
gaps_col = cumsum(c(5,5)))