Background / scenario:
I have two tables: One 1-2 million entry table with transactions of the form
TRX-ID , PROCESS-ID , ACTOR-ID
Additionally a participant-lookup (one of multiple categories of users of the system) table of the form
USER-ID , PARTICIPANT-ID
The transaction table is for historical reasons a bit messy. The PROCESS-ID can be a participant-id and the ACTOR-ID the user-id of a different kind of user. In some situations the PROCESS-ID is something else and the ACTOR-ID is the user-id of the participant.
I need to join the transaction and the participant-lookup table in order to get the participant-id for all transactions. I tried this in two ways.
(I left out some code steps in the snippets and focused on the join parts. Assume that df variables are data frames and the I select right columns to support e.g. unions.)
First approach:
transactions_df.join(pt_lookup_df, (transactions_df['actor-id'] == pt_lookup_df['user-id']) | (transactions_df['process-id'] == pt_lookup_df['participant-id']))
The code with this join is extrem slow. It ends up in my job running 45 minutes on a 10 instances AWS glue cluster with nearly 99% load on all executers.
Second approach:
I realised that some of the transactions already have the participant-id and I do not need to join for them. So I changed to:
transactions_df_1.join(pt_lookup_df, (transactions_df_1['actor-id'] == pt_lookup_df['user-id']))
transactions_df_2 = transactions_df_2.withColumnRenamed('process-id', 'participant-id')
transactions_df_1.union(transactions_df_2)
This finished in 5 minutes!
Both approaches give the correct results.
Question
I do not understand why the one is so slow and the other not. The amount of data excluded in the second approach is minimal. So transactions_df_2 has only a very small subset of the total data.
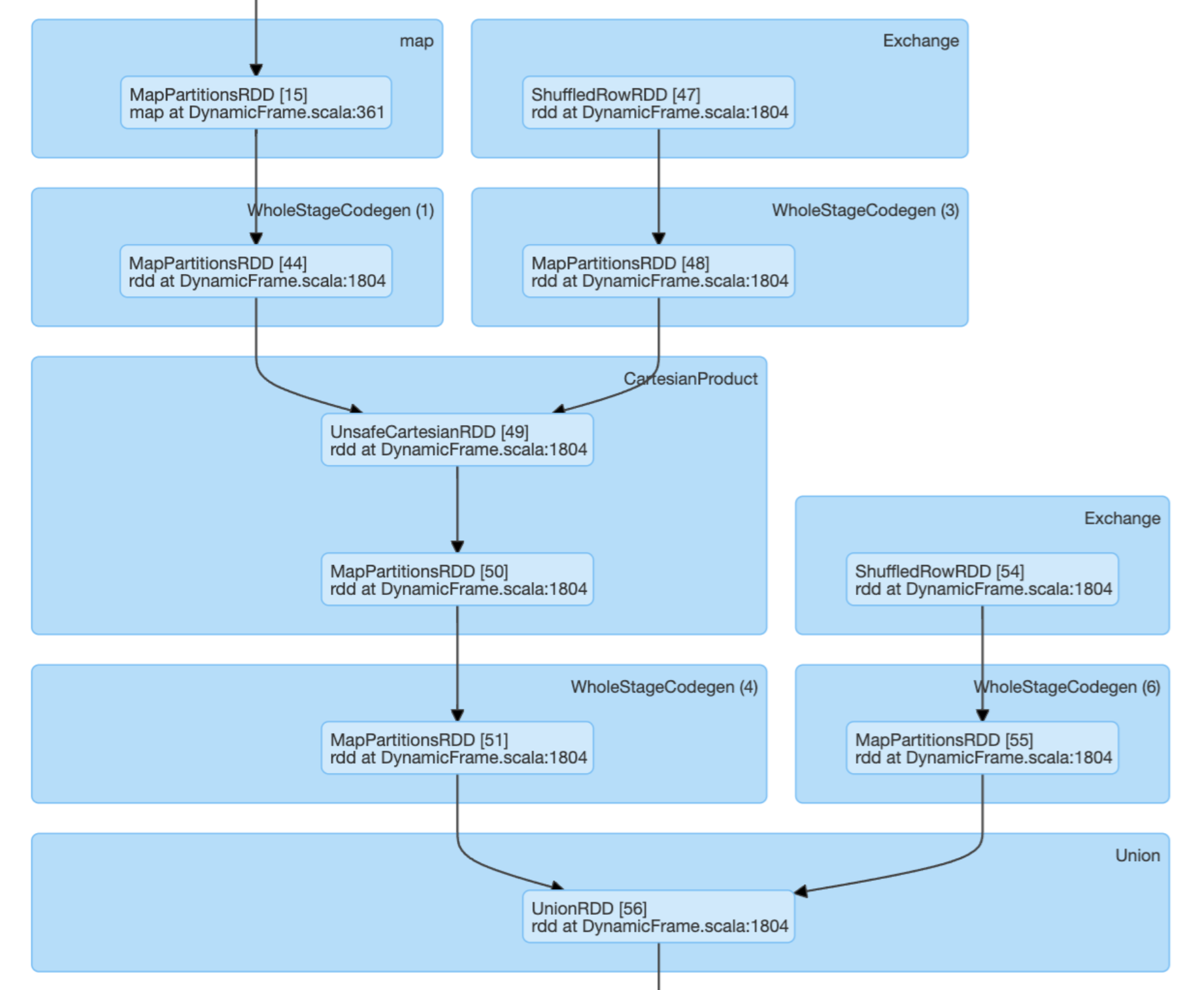
Looking at the plans, the affect is mainly around on Cartesian product that is done in approach 1 but not 2. So I assume, this is the performance bottleneck. I still do not understand how this can lead to 40 minutes differences in compute time.
Can someone give explanations?
Would a Cartesian product in the DAG be in general a warning sign in Spark?
Summary
It seems that a join with multiple columns in the condition triggers an extrem slow Cartesian product operation. Should I have done a broadcast operation on the smaller data set to avoid this?
DAG approach 1
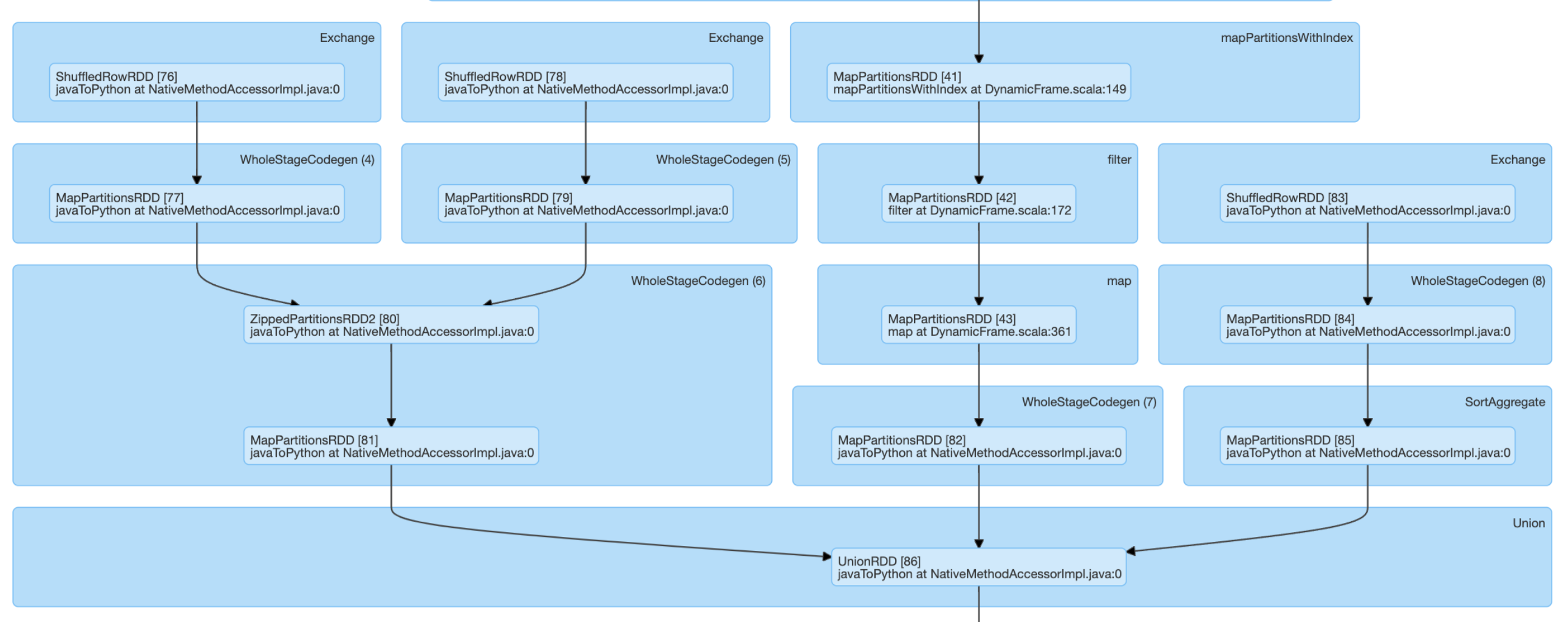
DAG approach 2
CodePudding user response:
This is because a Cartesian Product join and a regular join do not involve the same data shuffling process. Even thought the amount of data is similar the amount of shuffling is different.
This article explains where is the extra shuffling coming from.