Hello I have a simple problem I need to find specific lines in txt file they have to contain 'LG' which look like this:
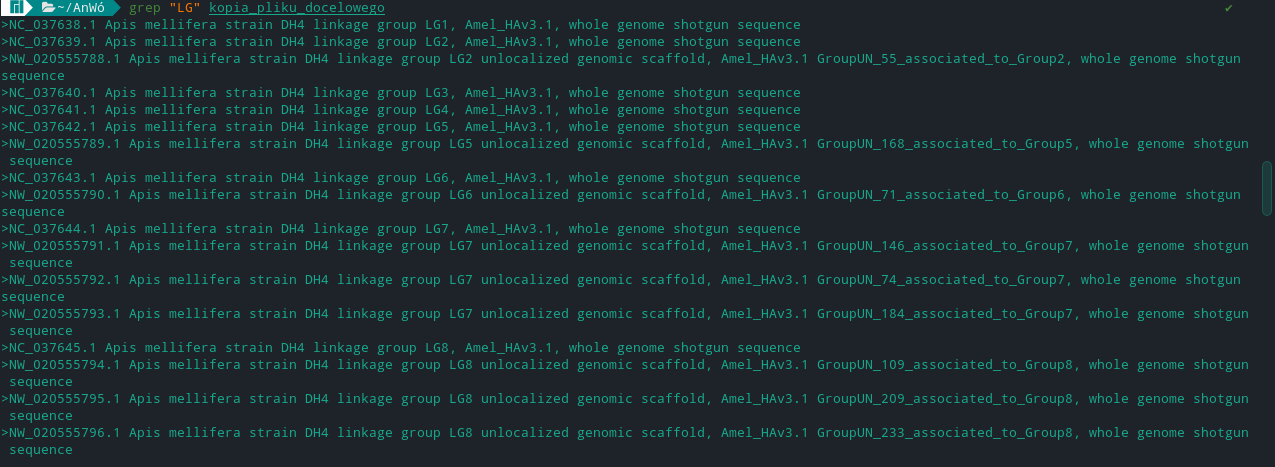
>NC_037638.1 Apis mellifera strain DH4 linkage group LG1, Amel_HAv3.1, whole genome shotgun sequence
then I need to replace number in this case NC_037638.1 with LG1
The LG and number will differ in each line
the result should look like this:
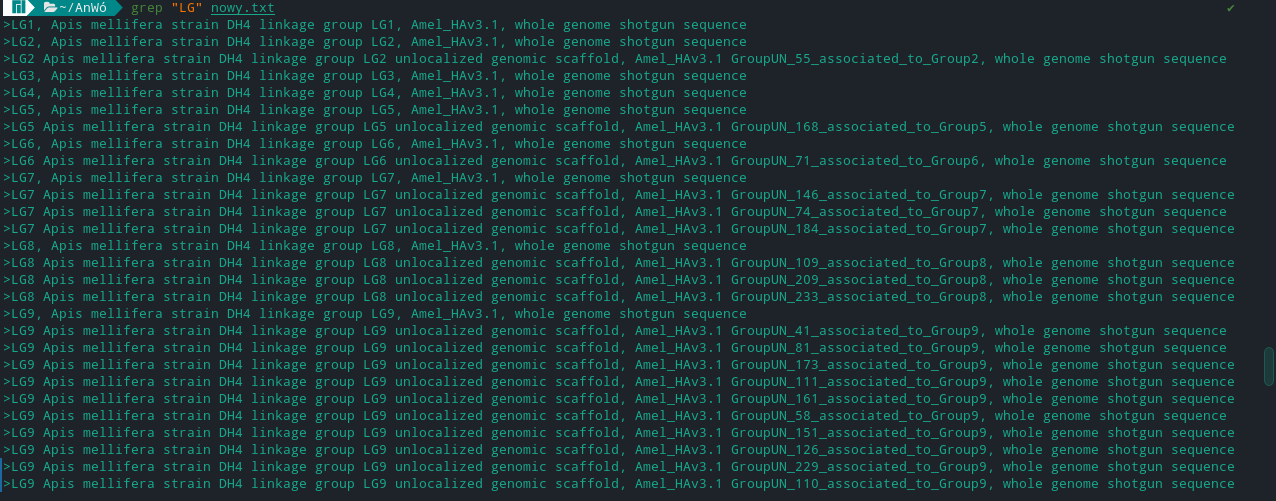
>LG1, Apis mellifera strain DH4 linkage group LG1, Amel_HAv3.1, whole genome shotgun sequence
I have like 3 mil of lines in a file and I need to find only those with LG followed by some number like in the example LG1
So basically i need to get from this:
I wrote something like this:
#!/bin/bash
while IFS= read -r line; do
if [[ $line =~ "LG" ]]; then
echo $line | awk ' { t = $1; $1 = $8; print; } ' | sed -e 's/^/>/' >> nowy.txt
else
echo $line >> nowy.txt
fi
done < kopia_pliku_docelowego
and it works but its ultra slow it takes like 3 minutes for the script to end
I thought out about solution and i figured i can grep for line index and change only those lines then swap old lines on the same index as new rewritten one.
I know how to find index (grep -n)
and i know how to change the line (talking about swaping number with LG)
but I don't know how to put it all together.
I would really appreciate some help
CodePudding user response:
I don't really understand the problem description. It sounds like you just want to replace the first column with the 8th column in any line that contains LG. If that's the case, just do:
awk '/LG/{ $1 = $8 }1' kopia_pliku_docelowego > nowy.txt
but perhaps you want to restrict the match so that you only do the replacements when 'LG' appears in the 8th column. You could do that with:
awk '$8 ~ /LG/{ $1 = $8 }1'
If you require that LG be followed by a string of digits, use:
awk '$8 ~ /LG[0-9] /{ $1 = $8 }1'
If you have lines in which the 8th column is LGxxxAAA (non string values following the digits) and you only want to replace the first column with that portion of the string that matches LG[0-9 ], you could use:
awk 'match($8,/LG[0-9] /){ $1 = substr($8,0,RLENGTH) }1'
awk can undoubtedly solve your problem, but you need to clarify exactly what you're trying to match. Your sed solution seems to be inserting a leading > which does not seem necessary according to your description. More specificity is required.
CodePudding user response:
You may do this in a single sed:
sed -i.bak -E 's/^>NC_037638\.1(.* (LG[0-9] ))/>\2\1/' file
cat file
>LG1 Apis mellifera strain DH4 linkage group LG1, Amel_HAv3.1, whole genome shotgun sequence
Explanation:
^>: Match>after start positionNC_037638\.1: Match textNC_037638.1(.*: Nn capture group #1 match & capture any text followed by a space followed by...(LG[0-9] )): MatchLGfollowed by 1 numbers in capture group #2>\2\1: Replacement part to have>followed byLGsubstring (what we captured in group #2) followed by back-reference of capture group #1
CodePudding user response:
Just awk, maybe:
awk '{
for(i=1;i<NF-1;i )
if($i=="linkage" && $(i 1)=="group")
break
if(i!=NF-1)
$1=$(i 2)
print
}' file.txt
We search for the two consecutive words "linkage" and "group", just in case they are not always at the same position in the lines. I suspect it could happen because of "Apis mellifera" that looks like a single field containing a space. If we find these two words we replace the first field with the one that follows "linkage group".
If the field following "linkage group" must be further constrained, e.g. to be LGnnn where nnn is some string of digits, we can change a bit the condition:
awk '{
for(i=1;i<NF-1;i )
if($i=="linkage" && $(i 1)=="group" && $(i 2) ~ /^LG[[:digit:]] /)
break;
if(i!=NF-1)
$1=$(i 2)
print
}' file.txt