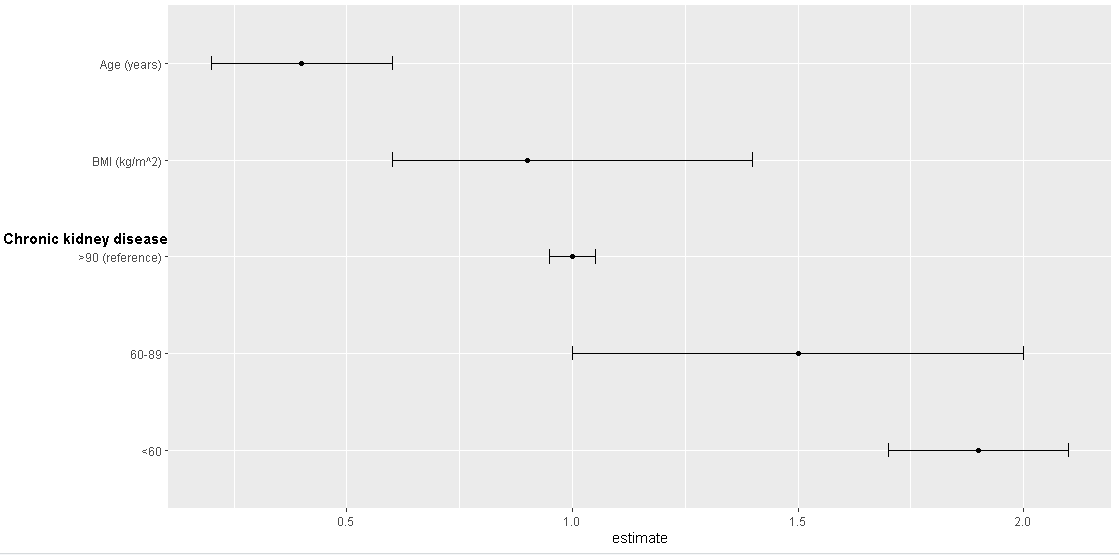
Ive got the estimates and corresponding 95% confidence intervals from a regression model, which I am trying to plot.
The data looks as follows:
library(tidyverse)
mydata <- structure(list(term = structure(c(1L, 7L, 18L, 19L, 20L), .Label = c("Age (years)",
"Sex (male)", "Never smoker (reference)", "Current smoker", "Former smoker",
"Obesity", "BMI (kg/m^2)", "Diabetes", "Glucose (mmol/L)", "Glucose lowering medication use",
"Hypertension", "Systolic blood pressure (mmHg)", "Diastolic blood pressure (mmHg)",
"Antihypertensive medication use", "Hypercholesterolemia", "LDL cholesterol (mmol/L)",
"Lipid lowering medication use", ">90 (reference)", "60-89",
"<60"), class = c("ordered", "factor")), estimate = c(0.4, 0.9,
1, 1.5, 1.9), conf_low = c(0.2, 1.4, 0.95, 1, 1.7), conf_high = c(0.6,
0.6, 1.05, 2, 2.1)), row.names = c(NA, -5L), class = c("tbl_df",
"tbl", "data.frame"))
mydata
# A tibble: 5 x 4
term estimate conf_low conf_high
<ord> <dbl> <dbl> <dbl>
1 Age (years) 0.4 0.2 0.6
2 BMI (kg/m^2) 0.9 1.4 0.6
3 >90 (reference) 1 0.95 1.05
4 60-89 1.5 1 2
5 <60 1.9 1.7 2.1
I have plotted these numbers as follows:
ggplot(data=mydata,
aes(x=estimate, y=fct_rev(term)))
geom_point()
geom_errorbarh(aes(xmin=conf_low, xmax=conf_high, height=0.15))
annotation_custom(grob=grid::textGrob(label="Chronic kidney disease",
gp=grid::gpar(fontface="bold", fontsize=11),
hjust=1.0),
xmin=-Inf, xmax=-Inf, ymin=3.2, ymax=3.2)
coord_cartesian(clip="off")
#scale_x_continuous(expand=expansion(mult=c(.05, .3)))
theme(axis.text.y=element_text(margin=margin(t=0, r=2.2, b=0, l=40, "pt")))
scale_y_discrete(name="")
I'd like to achieve the following things which I can't get to work:
In the label of BMI on the Y axis, the ^2 is supposed to be superscript. I've tried
expression()but that doesn't seem to work.In the labeling of the Chronic Kidney Disease variable on the y axis, the >90 is supposed to be a 'greater than or equal to' sign, and the <60 an 'smaller than or equal to'. Here I've tried expression and unicodes, but both don't seem to work.
Edit Added session info as this might be useful:
library(utils)
sessionInfo(package=NULL)
R version 4.1.2 (2021-11-01)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 7 x64 (build 7601) Service Pack 1
Matrix products: default
locale:
[1] LC_COLLATE=Dutch_Netherlands.1252 LC_CTYPE=Dutch_Netherlands.1252 LC_MONETARY=Dutch_Netherlands.1252
[4] LC_NUMERIC=C LC_TIME=Dutch_Netherlands.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_2.1.0 tidyr_1.1.4 tibble_3.1.6
[8] ggplot2_3.3.5 tidyverse_1.3.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 cellranger_1.1.0 pillar_1.6.4 compiler_4.1.2 dbplyr_2.1.1 tools_4.1.2
[7] digest_0.6.28 jsonlite_1.7.2 lubridate_1.8.0 lifecycle_1.0.1 gtable_0.3.0 pkgconfig_2.0.3
[13] rlang_0.4.12 reprex_2.0.1 cli_3.1.0 rstudioapi_0.13 DBI_1.1.1 haven_2.4.3
[19] xml2_1.3.2 withr_2.4.2 httr_1.4.2 fs_1.5.0 generics_0.1.1 vctrs_0.3.8
[25] hms_1.1.1 grid_4.1.2 tidyselect_1.1.1 glue_1.5.0 R6_2.5.1 fansi_0.5.0
[31] readxl_1.3.1 farver_2.1.0 tzdb_0.2.0 modelr_0.1.8 magrittr_2.0.1 backports_1.3.0
[37] scales_1.1.1 ellipsis_0.3.2 rvest_1.0.2 assertthat_0.2.1 colorspace_2.0-2 labeling_0.4.2
[43] utf8_1.2.2 stringi_1.7.5 munsell_0.5.0 broom_0.7.10 crayon_1.4.2
CodePudding user response:
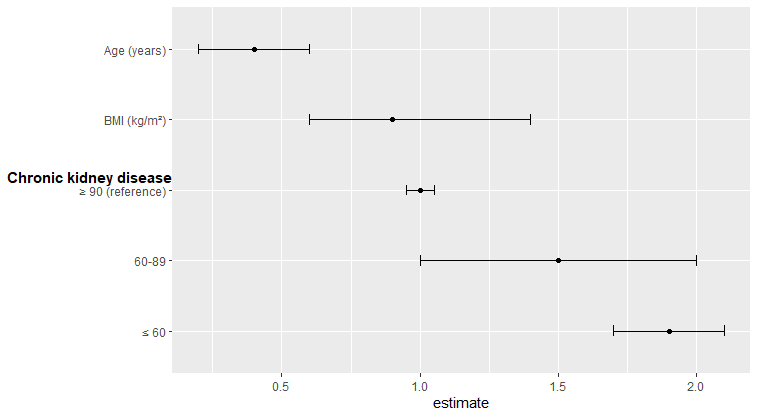
How did you add the unicode?
"BMI (kg/m\u00b2)"
"\u2265 90 (reference)" or "\u226590 (reference)"
"\u2264 60" or "\u226460"
This seem to work.