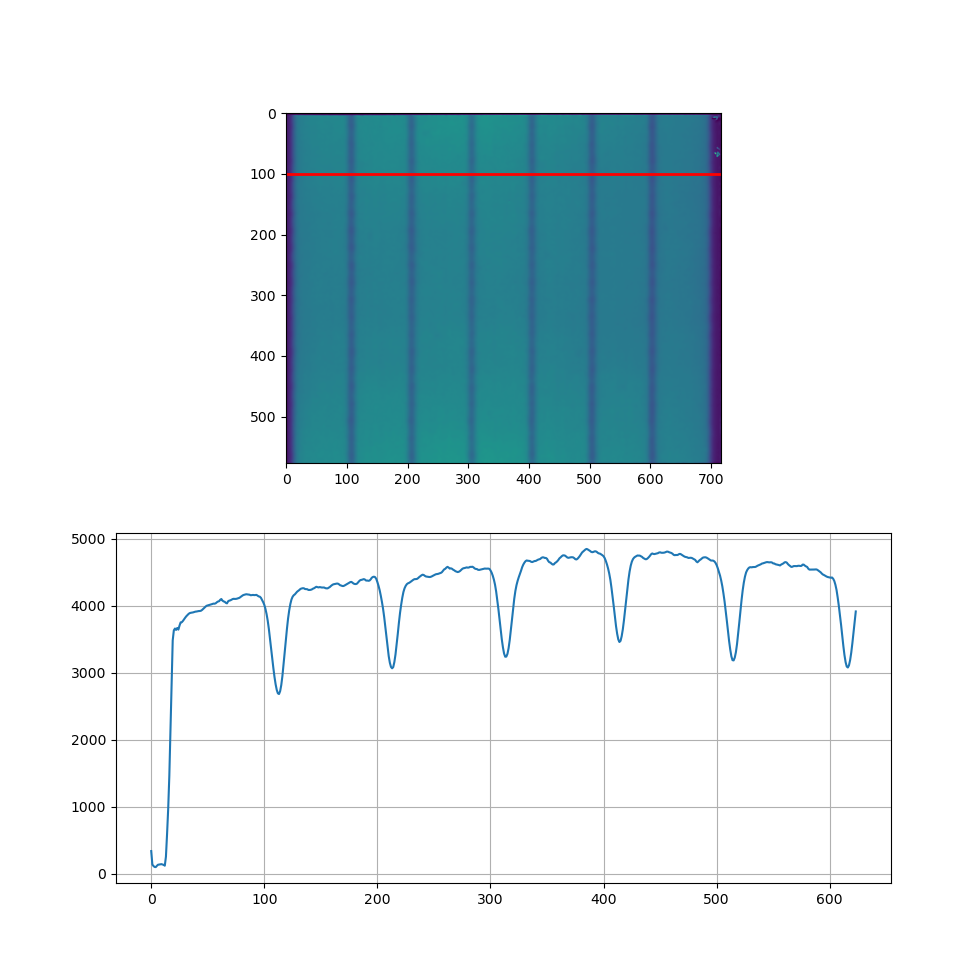
Why does the intensity profile of the image which I'm processing not include the right end? I'm supposed to see 7 "peaks" but I don't.
Below is my code:
from matplotlib import pyplot as plt
from skimage.measure import profile_line
from skimage.io import imread
start = (0, 714) #Start of the profile line
end = (100, 100) #End of the profile line
#image = imread('....tif') #Read the images
profile = profile_line(image, start, end, linewidth=1, mode='constant') #Take the profile line
fig, ax = plt.subplots(2, 1, figsize=(10, 10)) #Create the figures
ax[0].imshow(image) #Show the film at the top
ax[0].plot(start, end, 'r-', lw=2) #Plot a red line across the film
ax[1].plot(profile)
ax[1].grid()
Resulting figure:
The film which I'm processing:
CodePudding user response:
You are confusing a bunch of different concepts here. For one thing, your visualization is wrong. 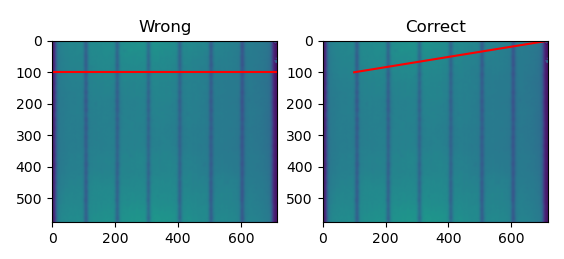
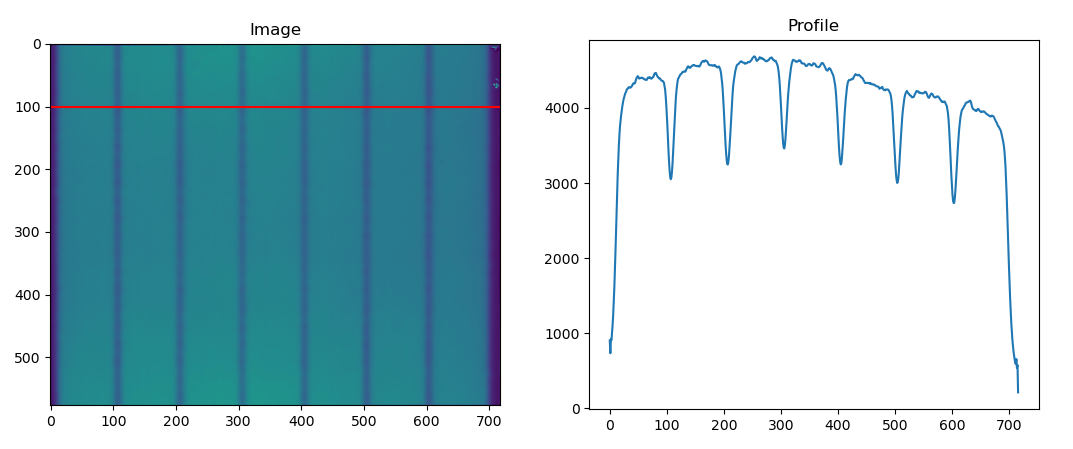
Hopefully you see that the image labeled "Correct" is showing exactly what you are seeing in your plot.
You can easily correct this. First, let's do it the complicated way: using profile_line. You want your start and end y-coordinates to be the same, and the start and end x-coordinates to span the image. Keep in mind that profile_line is inclusive on both ends:
image = skimage.io.imread(...)
start = (100, 0) #Start of the profile line row=100, col=0
end = (100, image.shape[1] - 1) #End of the profile line row=100, col=last
profile = skimage.measure.profile_line(image, start, end)
fig, ax = plt.subplots(1, 2)
ax[0].set_title('Image')
ax[0].imshow(image)
ax[0].plot([start[1], end[1]], [start[0], end[0]], 'r')
ax[1].set_title('Profile')
ax[1].plot(profile)
This looks much better. For vertical and horizontal cross-sections, you can obtain identical results using simple indexing. The images loaded by skimage.io.imread are numpy arrays, so you can just do:
profile = image[100, :]
For a vertical line, say at column 202, you index along the second (column) axis:
profile = image[:, 202]