This is probably an easy task for ppl more familiar with ggplot2 than I am. I have this type of data, increase_max grouped by role, which has two levels:
df <- structure(list(role = c("Recipient", "Speaker", "Recipient",
"Recipient", "Recipient", "Speaker", "Recipient", "Recipient",
"Speaker", "Speaker", "Recipient", "Speaker", "Recipient", "Recipient",
"Recipient", "Speaker", "Recipient", "Speaker", "Recipient",
"Speaker", "Recipient", "Recipient", "Speaker", "Recipient",
"Recipient", "Speaker", "Speaker", "Speaker", "Recipient", "Speaker",
"Speaker", "Recipient", "Speaker", "Recipient", "Recipient",
"Speaker", "Recipient", "Recipient", "Recipient", "Speaker",
"Speaker", "Recipient", "Speaker", "Recipient", "Speaker", "Recipient",
"Speaker", "Speaker", "Recipient", "Recipient", "Speaker", "Recipient",
"Recipient", "Speaker", "Recipient", "Recipient", "Recipient",
"Speaker", "Recipient", "Speaker", "Recipient", "Speaker", "Recipient",
"Recipient", "Speaker", "Recipient", "Recipient", "Speaker",
"Recipient", "Recipient", "Recipient", "Speaker", "Recipient",
"Speaker", "Recipient", "Speaker", "Recipient", "Recipient",
"Recipient", "Recipient", "Speaker", "Recipient", "Recipient",
"Recipient", "Speaker", "Recipient", "Speaker", "Recipient",
"Recipient", "Speaker", "Recipient", "Recipient", "Speaker",
"Recipient", "Recipient", "Recipient", "Speaker", "Recipient",
"Speaker", "Recipient"), increase_max = c(0.008, 0.118, NA, NA,
NA, 0.209, NA, 0.001, 0.111, NA, NA, NA, NA, NA, 0.007, 0.002,
0.006, 0.255, 0.009, NA, 0.004, 0.232, NA, 0.007, 0.004, 0.095,
0.09, NA, 0.002, NA, 0.05, NA, 0.02, 0.045, 0.002, NA, NA, 0.005,
0.012, NA, 0.037, NA, 0.066, NA, 0.019, 0.002, 0.136, NA, 0.003,
NA, 0.128, 0.004, 0.003, NA, NA, NA, 0.03, 0.042, NA, 0.138,
0.139, 0.126, 0.002, NA, 0.005, NA, 0.002, 0.01, 0.001, NA, 0.005,
0.003, NA, NA, 0.002, NA, 0.005, NA, NA, 0.015, 0.007, 0.021,
NA, NA, NA, NA, NA, 0.171, 0.02, 0.036, 0.026, 0.001, 0.033,
0.127, 0.339, 0.075, 0.037, 0.083, NA, 0.041)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -100L))
My way of producing the plot works, at least basically, but is surely utterly klunky and complicated:
# variable 1:
speaker_0 <- df %>%
filter(!is.na(increase_max)
& role == "Speaker") %>%
pull(increase_max)
# variable 2:
recipient_0 <- df %>%
filter(!is.na(increase_max)
& role == "Recipient") %>%
pull(increase_max)
# subset both variables on certain range:
speaker <- data.frame(Max_EDA_increase = speaker_0[speaker_0 >= 0.05 & speaker_0 <= 0.5])
recipient <- data.frame(Max_EDA_increase = recipient_0[recipient_0 >= 0.05 & recipient_0 <= 0.5])
# bind together:
both <- rbind(speaker, recipient)
# plot histogram with density lines:
ggplot(both, aes(x = Max_EDA_increase))
geom_histogram(aes(y = after_stat(density)), data = speaker, fill = "red", alpha = 0.35, binwidth = 0.05)
geom_line(data = speaker, color = "red", stat = "density", alpha = 0.35)
geom_histogram(aes(y = after_stat(density)), data = recipient, fill = "blue", alpha = 0.35, binwidth = 0.05)
geom_line(data = recipient, color = "blue", stat = "density", alpha = 0.35)
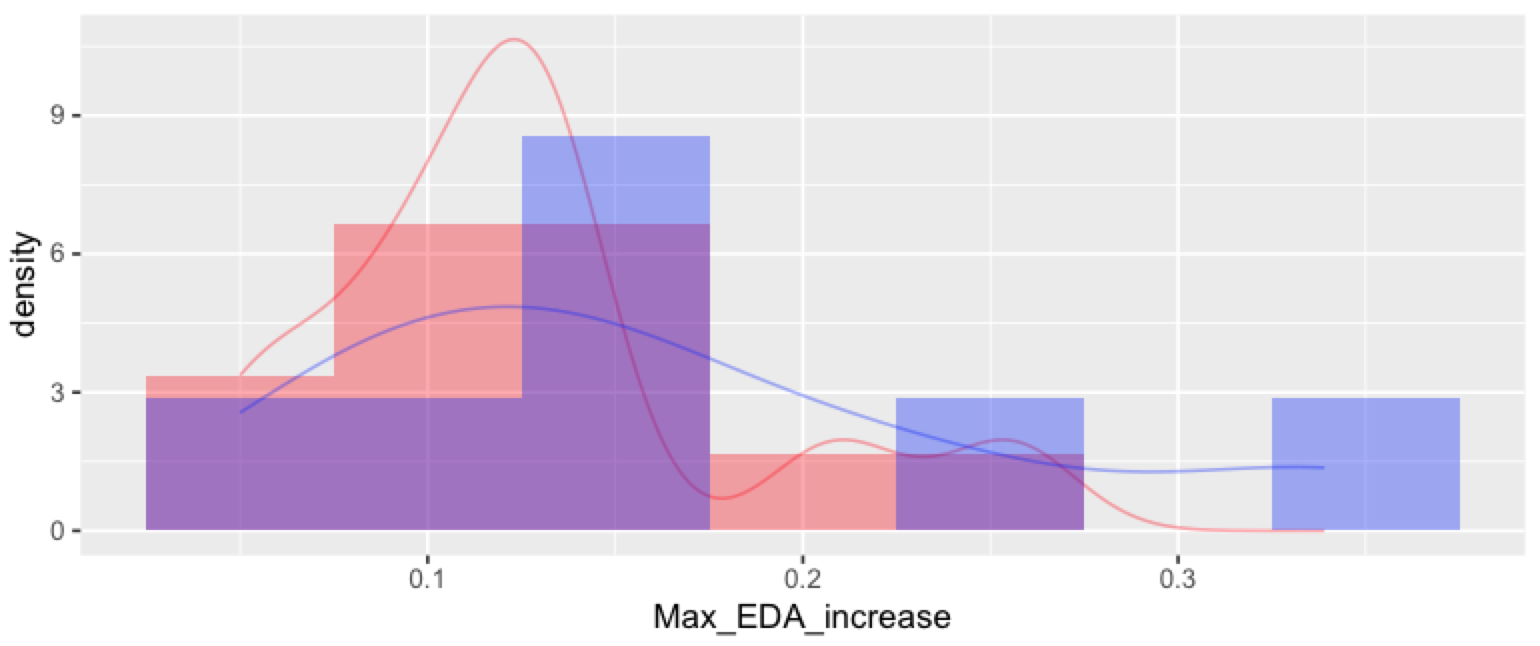
The resulting plot:
I'm certain there must be a more direct way to produce the plot, which also adds a legend to distinguish the two groups/two density lines!
CodePudding user response:
I think the way to make this less clunky is to not split-combine by role. You can filter the data once, and subsequently set fill = role or colour = role.
library(ggplot2)
# Omitted for brevity
# df <- structure(...)
df2 <- subset(df, !is.na(increase_max) &
increase_max >= 0.05 &
increase_max <= 0.5)
ggplot(df2, aes(x = increase_max))
geom_histogram(aes(y = after_stat(density), fill = role),
binwidth = 0.05, position = "identity",
alpha = 0.35)
geom_density(aes(colour = role))
scale_colour_manual(
aesthetics = c("fill", "colour"),
values = c("blue", "red")
)
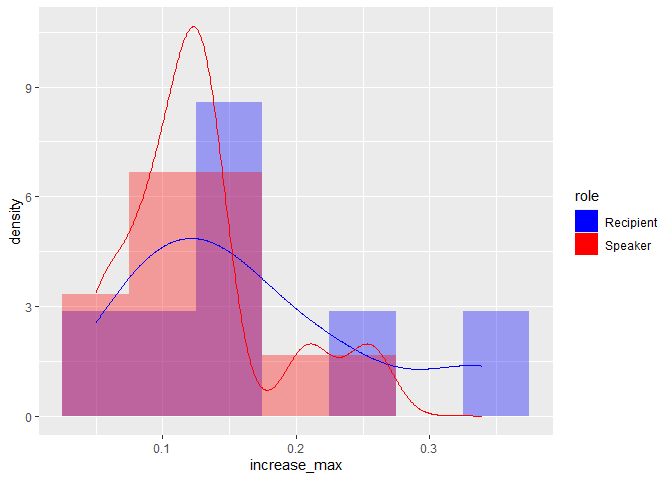
Created on 2021-12-14 by the reprex package (v2.0.1)