I have a data like that looks like the following:
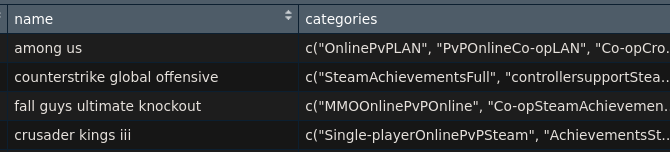
dput(head(my.data[c("name", "categories")],1))
structure(list(name = "among us", categories = list(c("OnlinePvPLAN",
"PvPOnlineCo-opLAN", "Co-opCross-PlatformMultiplayerRemote",
"Playon", "PhoneRemotePlay", "onTablet]"))), row.names = 1L, class = "data.frame")
As you can imagine this structure is not easy to deal with. I want to separate the categories column to multiple logical columns like the following:
name | OnlinePvPLAN | PvPOnlineCo-opLAN | MMOOnlinePvPOnline
-----|--------------|-------------------|-------------------- .....
among| TRUE | TRUE | FALSE
us
Since there are there are lot's of category columns I decided to write a function to write function that takes a list of categories to separate.
With the following code I can find columns that are in a particular category:
filter(my.data, map_lgl(my.data$categories, ~"OnlinePvPLAN" %in% .))
Using this, I wrote the following function:
compile.category.func <- function(data, category.list){
lapply(X=category.list, function(category){
category <- c(category)
mutate(data, category=ifelse(map_lgl(data$categories, ~category %in% .), TRUE, FALSE))
})
data
}
output <- compile.category.func(my.data, c("OnlinePvPLAN","MMOOnlinePvPOnline"))
However this function doesn't work and doesn't generate any new columns.
CodePudding user response:
unnest into long form, use table to create the frequencies, use mutate to convert to TRUE/FALSE, then as.data.frame.matrix to convert that to a data frame and finally add the name column back. Omit the mutate line if 1/0 is wanted instead.
library(dplyr)
library(tidyr)
library(tibble)
DF %>%
unnest_longer(categories) %>%
table %>%
as.data.frame.matrix %>%
mutate(across(, as.logical)) %>%
rownames_to_column("name")
giving
name Co-opCross-PlatformMultiplayerRemote OnlinePvPLAN onTablet]
1 among us TRUE TRUE TRUE
PhoneRemotePlay Playon PvPOnlineCo-opLAN
1 TRUE TRUE TRUE
