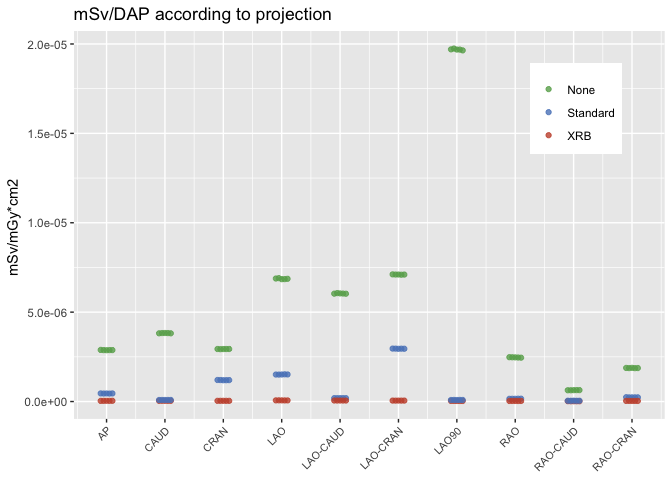
I have measurement from three groups that I would like to plot, each with five observations. I would like to plot all the points, however the numeric values are very close within each group. Changing alpha helps, but it is still difficult to observe the seperate points.
Thus I would like to add some horizontal spread (on theX-axis). Partial overlap of the points is ok. geom_jitter() can achieve this, but adds a random spread, whereas I would like to spread the five points uniformly in the X-axis as there is no randomness in the data in the horizontal axis.
Geom_dotplot() could create an even spread, but to my understanding only works with discrete / binned values.
Here is my ggplot output, and reproducible code with the data in tribble format and the ggplot code :
library(tidyverse)
df <- tribble(
~projection, ~E.DAP, ~Shielding,
"AP", 2.88755980861244e-06, "None",
"AP", 2.87626262626263e-06, "None",
"AP", 2.87e-06, "None",
"AP", 2.87702265372168e-06, "None",
"AP", 2.87775551102204e-06, "None",
"CRAN", 2.93893129770992e-06, "None",
"CRAN", 2.92857142857143e-06, "None",
"CRAN", 2.93517017828201e-06, "None",
"CRAN", 2.9375e-06, "None",
"CRAN", 2.93831168831169e-06, "None",
"LAO", 6.88053097345133e-06, "None",
"LAO", 6.90217391304348e-06, "None",
"LAO", 6.84489795918367e-06, "None",
"LAO", 6.84792626728111e-06, "None",
"LAO", 6.86425339366516e-06, "None",
"LAO-CRAN", 7.11216730038023e-06, "None",
"LAO-CRAN", 7.1039501039501e-06, "None",
"LAO-CRAN", 7.1042471042471e-06, "None",
"LAO-CRAN", 7.09368635437882e-06, "None",
"LAO-CRAN", 7.10183299389002e-06, "None",
"RAO", 2.48e-06, "None",
"RAO", 2.47540983606557e-06, "None",
"RAO", 2.46979865771812e-06, "None",
"RAO", 2.46206896551724e-06, "None",
"RAO", 2.453125e-06, "None",
"RAO-CRAN", 1.87972508591065e-06, "None",
"RAO-CRAN", 1.87068965517241e-06, "None",
"RAO-CRAN", 1.88461538461538e-06, "None",
"RAO-CRAN", 1.86816720257235e-06, "None",
"RAO-CRAN", 1.8735632183908e-06, "None",
"LAO-CAUD", 6.03324808184143e-06, "None",
"LAO-CAUD", 6.06502242152466e-06, "None",
"LAO-CAUD", 6.04785894206549e-06, "None",
"LAO-CAUD", 6.0402144772118e-06, "None",
"LAO-CAUD", 6.02949061662198e-06, "None",
"RAO-CAUD", 6.30573248407643e-07, "None",
"RAO-CAUD", 6.29834254143646e-07, "None",
"RAO-CAUD", 6.35467980295566e-07, "None",
"RAO-CAUD", 6.38297872340426e-07, "None",
"RAO-CAUD", 6.37755102040816e-07, "None",
"CAUD", 3.81597222222222e-06, "None",
"CAUD", 3.83083511777302e-06, "None",
"CAUD", 3.8330550918197e-06, "None",
"CAUD", 3.82936507936508e-06, "None",
"CAUD", 3.81614349775785e-06, "None",
"AP", 4.53580901856764e-07, "Standard",
"AP", 4.46745562130178e-07, "Standard",
"AP", 4.4973544973545e-07, "Standard",
"AP", 4.44976076555024e-07, "Standard",
"AP", 4.52380952380952e-07, "Standard",
"CRAN", 1.20574162679426e-06, "Standard",
"CRAN", 1.20113314447592e-06, "Standard",
"CRAN", 1.19130434782609e-06, "Standard",
"CRAN", 1.19349593495935e-06, "Standard",
"CRAN", 1.19757575757576e-06, "Standard",
"LAO", 1.50961538461538e-06, "Standard",
"LAO", 1.50761421319797e-06, "Standard",
"LAO", 1.51209677419355e-06, "Standard",
"LAO", 1.52216748768473e-06, "Standard",
"LAO", 1.51476793248945e-06, "Standard",
"LAO-CRAN", 2.96213425129088e-06, "Standard",
"LAO-CRAN", 2.95991561181435e-06, "Standard",
"LAO-CRAN", 2.95e-06, "Standard",
"LAO-CRAN", 2.95744680851064e-06, "Standard",
"LAO-CRAN", 2.95266272189349e-06, "Standard",
"RAO", 1.51515151515152e-07, "Standard",
"RAO", 1.52173913043478e-07, "Standard",
"RAO", 1.52866242038217e-07, "Standard",
"RAO", 1.57575757575758e-07, "Standard",
"RAO", 1.59420289855072e-07, "Standard",
"RAO-CRAN", 2.41379310344828e-07, "Standard",
"RAO-CRAN", 2.33050847457627e-07, "Standard",
"RAO-CRAN", 2.34741784037559e-07, "Standard",
"RAO-CRAN", 2.33812949640288e-07, "Standard",
"RAO-CRAN", 2.34817813765182e-07, "Standard",
"LAO-CAUD", 1.89125295508274e-07, "Standard",
"LAO-CAUD", 1.87110187110187e-07, "Standard",
"LAO-CAUD", 1.89309576837416e-07, "Standard",
"LAO-CAUD", 1.90821256038647e-07, "Standard",
"LAO-CAUD", 1.89189189189189e-07, "Standard",
"AP", 3.7542662116041e-08, "XRB",
"AP", 3.89972144846797e-08, "XRB",
"AP", 4.15335463258786e-08, "XRB",
"AP", 3.83275261324042e-08, "XRB",
"AP", 4.09556313993174e-08, "XRB",
"CRAN", 4e-08, "XRB",
"CRAN", 4.08163265306122e-08, "XRB",
"CRAN", 4.06905055487053e-08, "XRB",
"CRAN", 3.94574599260173e-08, "XRB",
"CRAN", 3.90835579514825e-08, "XRB",
"LAO", 6.47249190938511e-08, "XRB",
"LAO", 6.69144981412639e-08, "XRB",
"LAO", 6.42570281124498e-08, "XRB",
"LAO", 6.19834710743802e-08, "XRB",
"LAO", 6.25e-08, "XRB",
"LAO-CRAN", 5.31914893617021e-08, "XRB",
"LAO-CRAN", 5.11247443762781e-08, "XRB",
"LAO-CRAN", 5.30821917808219e-08, "XRB",
"LAO-CRAN", 5.17857142857143e-08, "XRB",
"LAO-CRAN", 5.31732418524871e-08, "XRB",
"RAO", 3.30578512396694e-08, "XRB",
"RAO", 3.38983050847458e-08, "XRB",
"RAO", 3.33333333333333e-08, "XRB",
"RAO", 3.07017543859649e-08, "XRB",
"RAO", 3.2171581769437e-08, "XRB",
"RAO-CRAN", 3.44827586206897e-08, "XRB",
"RAO-CRAN", 3.5264483627204e-08, "XRB",
"RAO-CRAN", 3.25581395348837e-08, "XRB",
"RAO-CRAN", 3.43007915567282e-08, "XRB",
"RAO-CRAN", 3.37837837837838e-08, "XRB",
"LAO-CAUD", 5.70776255707763e-08, "XRB",
"LAO-CAUD", 5.50847457627119e-08, "XRB",
"LAO-CAUD", 5.51876379690949e-08, "XRB",
"LAO-CAUD", 5.42797494780793e-08, "XRB",
"LAO-CAUD", 5.53191489361702e-08, "XRB",
"RAO-CAUD", 2.1505376344086e-08, "XRB",
"RAO-CAUD", 1.52990264255911e-08, "XRB",
"RAO-CAUD", 1.58033362598771e-08, "XRB",
"RAO-CAUD", 1.47679324894515e-08, "XRB",
"RAO-CAUD", 1.60213618157543e-08, "XRB",
"CAUD", 3.84024577572965e-08, "XRB",
"CAUD", 3.98671096345515e-08, "XRB",
"CAUD", 4e-08, "XRB",
"CAUD", 3.92156862745098e-08, "XRB",
"CAUD", 3.94574599260173e-08, "XRB",
"RAO-CAUD", 4.52961672473868e-08, "Standard",
"RAO-CAUD", 4.12844036697248e-08, "Standard",
"RAO-CAUD", 4.83870967741936e-08, "Standard",
"RAO-CAUD", 4.29184549356223e-08, "Standard",
"RAO-CAUD", 4.38356164383562e-08, "Standard",
"CAUD", 8.58974358974359e-08, "Standard",
"CAUD", 8.70646766169154e-08, "Standard",
"CAUD", 8.66372980910426e-08, "Standard",
"CAUD", 8.67579908675799e-08, "Standard",
"CAUD", 8.70113493064313e-08, "Standard",
"LAO90", 3.38266384778013e-08, "XRB",
"LAO90", 3.34346504559271e-08, "XRB",
"LAO90", 3.2258064516129e-08, "XRB",
"LAO90", 3.21543408360129e-08, "XRB",
"LAO90", 3.20987654320988e-08, "XRB",
"LAO90", 8.60215053763441e-08, "Standard",
"LAO90", 8.88888888888889e-08, "Standard",
"LAO90", 8.44686648501362e-08, "Standard",
"LAO90", 8.90804597701149e-08, "Standard",
"LAO90", 8.67052023121387e-08, "Standard",
"LAO90", 1.96982378854626e-05, "None",
"LAO90", 1.97443820224719e-05, "None",
"LAO90", 1.96883720930233e-05, "None",
"LAO90", 1.96856540084388e-05, "None",
"LAO90", 1.96415770609319e-05, "None"
)
ggplot(df , aes(x=projection, y=E.DAP, color=Shielding))
geom_jitter(size = 2.5, width=0.2, height=0, alpha =0.8)
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1, size=8))
labs(title= "mSv/DAP according to projection",
y="mSv/mGy*cm2 ", x="")
theme(legend.position = c(0.85, 0.8), legend.key = element_rect(colour = "transparent", fill = "transparent"))
scale_color_manual(values=c(c("#69ac5c", "#5b86c3","#c7533b")))
theme(legend.title = element_blank())
CodePudding user response:
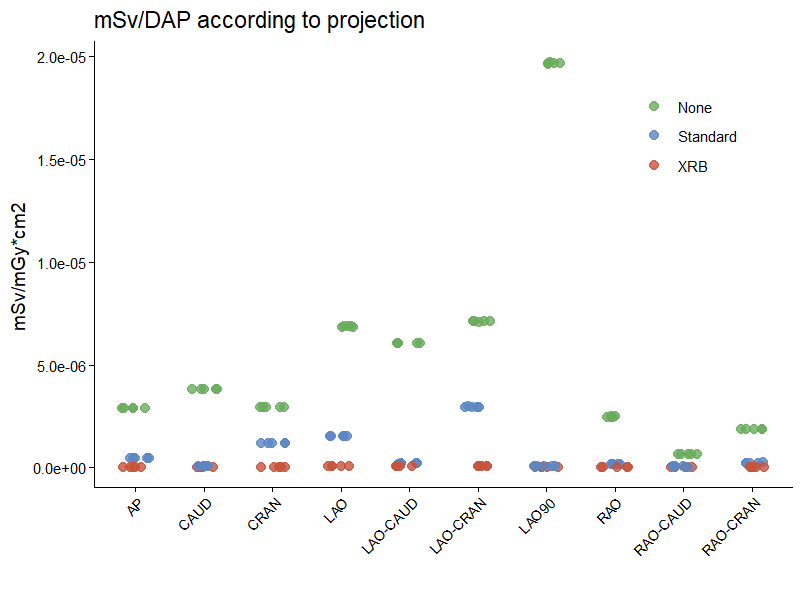
Package ggbeeswarm could be used...
The appearance could be enhanced with a different shape or the arguments of geom_beeswarm: alpha, dodge.width or groupOnX.
Setting dodge.width forces the points to be grouped by the Shielding variable. The updated answer, nominally includes a value of 0.1, setting dodge.width closer to zero makes is seem as if there is no dodging and keeps points from the same Shielding value together. A larger dodge.width value will help distinguish points for x variables such as "CAUD".
To ensure overplotting by Shielding is consistent create a plotting order variable. Arbitrarily set by arranging the data by Shielding so that the last variable plots last, this could be altered to suit.
library(ggplot2)
library(dplyr)
library(ggbeeswarm)
df1 <-
df %>%
arrange(Shielding) %>%
mutate(plot_order = row_number())
ggplot(df1, aes(x = projection, y = E.DAP, color = Shielding, order = plot_order))
geom_beeswarm(size = 2.5, alpha = 0.9, dodge.width = 0.1)
scale_color_manual(values=c(c("#69ac5c", "#5b86c3","#c7533b")))
labs(title= "mSv/DAP according to projection",
y="mSv/mGy*cm2 ", x="")
theme(legend.title = element_blank(),
legend.position = c(0.85, 0.8),
legend.key = element_rect(colour = "transparent", fill = "transparent"),
axis.text.x = element_text(angle = 45, vjust = 1, hjust=1, size=8))
Created on 2021-12-21 by the reprex package (v2.0.1)
CodePudding user response:
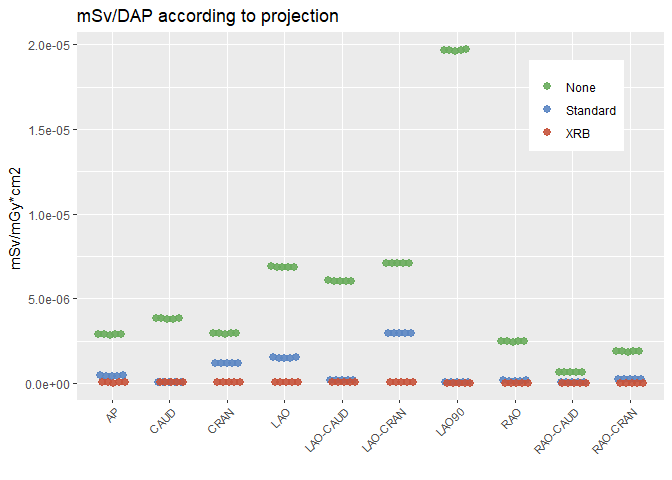
You can manually calculate an offset after transforming your x into a continuous variable.
library(tidyverse)
## prepare x
df <- df %>%
group_by(projection) %>%
mutate(
group = cur_group_id(),
x = rep(seq(unique(group) - .1, unique(group) .1, 0.05), 3)
)
ggplot(df, aes(x = x, y = E.DAP, color = Shielding))
## you can use geom_point now
geom_point(alpha = 0.8)
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1, size = 8))
labs(title = "mSv/DAP according to projection",
## use x = NULL and color = NULL instead of your theme element
y = "mSv/mGy*cm2 ", x = NULL, color = NULL
)
theme(legend.position = c(0.85, 0.8), legend.key = element_rect(colour = "transparent", fill = "transparent"))
scale_color_manual(values = c(c("#69ac5c", "#5b86c3", "#c7533b")))
## you will now need to set the breaks and labels
scale_x_continuous(breaks = seq_len(max(df$group)), labels = sort(unique(df$projection)))