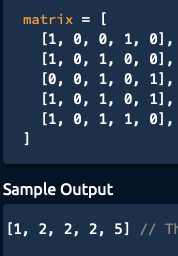
I am given a matrix containing only 1's and 0's, where adjacent 1's (horizontal or vertical, NOT diagonal) form rivers. I am asked to return an array of all river sizes in the matrix.
(Note the order of the river sizes don't matter- they can be in any order) i.e. [2,1,2,2,5] is also a valid solution!
I am writing the recursive part of the code, where as we explore the matrix, if we stumble on a 1, we will run a recursion to explore its adjacent left, right,up and down for any 1's.
The code is quite lengthy, and I don't believe it is necessary for my question. My question is, as I call on the recursive function, and I update currentRiverSize 1 (initialised as 0) in the argument of the recursive function as we stumble across more 1's, can I then at the end simply append this to the array?
I have a feeling the recursion is not structured correctly!
def findAllRiverSizes(matrix):
riverSizes = []
exploreRiver(matrix,row,col,entryExplored,riverSizes,0):
def exploreRiver(matrix,row,col,entryExplored,riverSizes,currentRiverSize):
#look right:
if col<len(matrix[0])-1 and not entryExplored[row][col 1] and matrix[row][col 1] == 1:
entryExplored[row][col 1] = True
exploreRiver(matrix,row,col 1,entryExplored,riverSizes,currentRiverSize 1)
#look left
if col>0 and not entryExplored[row][col-1] and matrix[row][col-1] == 1:
entryExplored[row][col-1] = True
exploreRiver(matrix,row,col-1,entryExplored,riverSizes,currentRiverSize 1)
#look up
if row >0 and not entryExplored[row-1][col] and matrix[row-1][col] == 1:
entryExplored[row-1][col] = True
exploreRiver(matrix,row-1,col,entryExplored,riverSizes,currentRiverSize 1)
#look down
if row <len(matrix)-1 and not entryExplored[row 1][col] and matrix[row 1][col] == 1:
entryExplored[row 1][col] = True
exploreRiver(matrix,row 1,col,entryExplored,riverSizes,currentRiverSize 1)
riverSizes.append(currentRiverSize)
return riverSizes
CodePudding user response:
The structure of your code doesn't seem to be doing what you describe you want it to be doing. Some suggestions -
- Instead of using the 4 different conditions try to use a single condition at the top which checks for the bounds
- Are we allowed to modify the matrix? In that case we can do away without the
entryExplored - We don't need to pass the
riverSizesto the DFS recursion function(since we only need to append the value toriverSizesonce per DFS run), so we can make the function return that value instead.
Here's a simplified code that does what you want -
def findAllRiverSizes(matrix):
def exploreRiverDFS(matrix,row,col):
# go through all neighbors of(row,col) having value 1,
# set the value to 0(or use entryExplored). In the end after
# there are no neighbors append the currentRiverSize to riverSizes
# return the size of this river
# check if current row, col is valid or there is no river in the current cell
if (row < 0 or row >= len(matrix) or col < 0 or col >= len(matrix[row])
or matrix[row][col] == 0):
return 0
matrix[row][col] = 0
return 1 sum([exploreRiverDFS(matrix, row 1, col),
exploreRiverDFS(matrix, row-1, col),
exploreRiverDFS(matrix, row, col 1),
exploreRiverDFS(matrix, row, col-1)])
riverSizes = []
# iterate through our matrix here, if we come across a 1, call the DFS function
# to go through all its neighboring cells and set them to 0 in matrix itself
# if you are allowed to modify matrix(else you can use entryExplored matrix)
# the exploreRiverDFS function returns the size of current river it traversed through
for row in range(len(matrix)):
for col in range(len(matrix[row])):
if matrix[row][col]:
riverSizes.append(exploreRiverDFS(matrix, row, col))
return riverSizes