GOAL
I have values v given at specific 3D coordinates x y z. The data is stored as a pandas dataframe:
x y z v
0 -68.5 68.50 -10.00 0.297845
1 -68.5 -23.29 61.10 0.148683
2 -68.5 -23.29 63.47 0.142325
3 -68.5 -23.29 65.84 0.135908
4 -68.5 -23.29 68.21 0.129365
... ... ... ...
91804 68.5 23.29 151.16 0.118460
91805 68.5 23.29 153.53 0.119462
91806 68.5 23.29 155.90 0.120386
91807 68.5 23.29 139.31 0.112257
91808 68.5 -68.50 227.00 0.127948
I would like to find the values at new coordinates that are not part of the dataframe, hence I am looking into how to efficiently interpolate the data.
What I have done:
Since the coordinates are on a grid, I can use 
I am wondering, what I am doing wrong??
Other:
Dataset
Please find the complete dataset here: https://filebin.net/u10lrw956enqhg5i
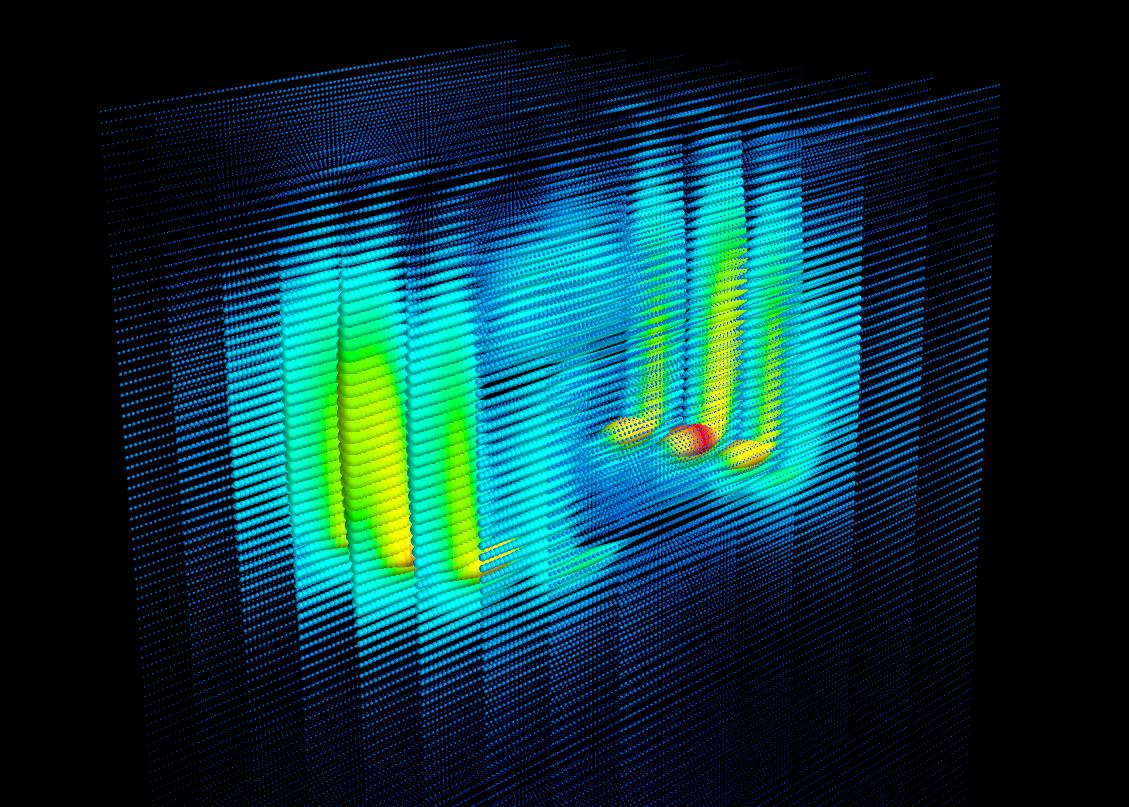
Visualization
from mayavi import mlab
# Create figure
fig = mlab.figure(1, fgcolor=(0, 0, 0), bgcolor=(0, 0, 0))
mlab.points3d(xs,ys,zs,output)
mlab.view(azimuth=270, elevation=90, roll=180, figure=fig)
# View plot
mlab.show()
CodePudding user response:
I strongly suspect that your data, while on a grid, is not ordered so as to allow a simple reshape of the values. You have two solutions available, both involving reordering the data in different ways.
Solution 1
Since you're already using np.unique to extract the grid, you can get the correct ordering of vs using the return_inverse parameter:
px, ix = np.unique(xs, return_inverse=True)
py, iy = np.unique(ys, return_inverse=True)
pz, iz = np.unique(zs, return_inverse=True)
points = (px, py, pz)
values = np.empty_like(vs, shape=(px.size, py.size, pz.size))
values[ix, iy, iz] = vs
return_inverse is sort of magical, largely because it's so counterintuitive. In this case, for each element of values, it tells you which unique, sorted gross location it corresponds to.
By the way, if you are missing grid elements, you may want to replace np.empty_like(vs, shape=(px.size, py.size, pz.size)) with either np.zeros_like(vs, shape=(px.size, py.size, pz.size)) or np.empty_like(vs, np.nan, shape=(px.size, py.size, pz.size)). In the latter case, you could interpolate the nans in the grid first.
Solution 2
The more obvious solution would be to rearrange the indices so you can reshape vs as you tried to do. That only works if you're sure that there are no missing grid elements. The easiest way would be to sort the whole dataframe, since the pandas methods are less annoying than np.lexsort (IMO):
df.sort_values(['x', 'y', 'z'], inplace=True, ignore_index=True)
When you extract, do it efficiently:
xs, ys, zs, vs = df.to_numpy().T
Since everything is sorted, you don't need np.unique to identify the grid any more. The number of unique x values is:
nx = np.count_nonzero(np.diff(xs)) 1
And the unique values are:
bx = xs.size // nx
ux = xs[::bx]
y values go through a full cycle every bx elements, so
ny = np.count_nonzero(np.diff(ys[:bx])) 1
by = bx // ny
uy = ys[:bx:by]
And for z (bz == 1):
nz = np.count_nonzero(np.diff(zs[:by])) 1
uz = zs[:by]
Now you can construct your original arrays:
points = (ux, uy, uz)
values = vs.reshape(nx, ny, nz)