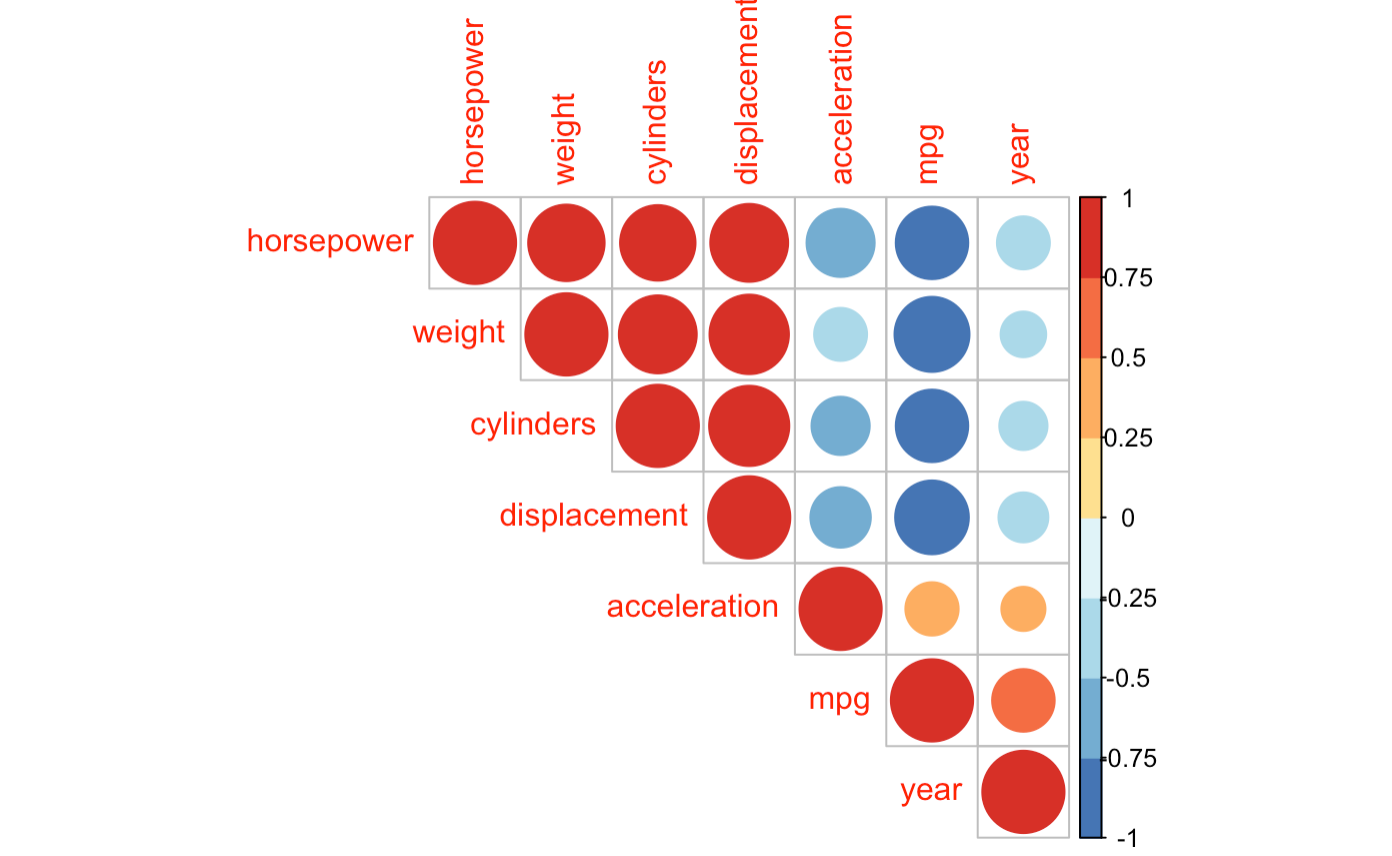
I created a correlation plot for my dataset in R but I am not sure how to choose which of the following pairs of variables express multicollinearity? An explanation with examples would be really helpful!
CodePudding user response:
Perhaps one way is through a qgraph. First I'll load the Holzinger data from the lavaan package, the correlation function from the correlation package, and the qgraph function with the qgraph package with the following libraries:
library(correlation)
library(qgraph)
library(lavaan)
Create the correlation matrix from the Holzinger data:
cor_holz <- HolzingerSwineford1939 %>%
correlation()
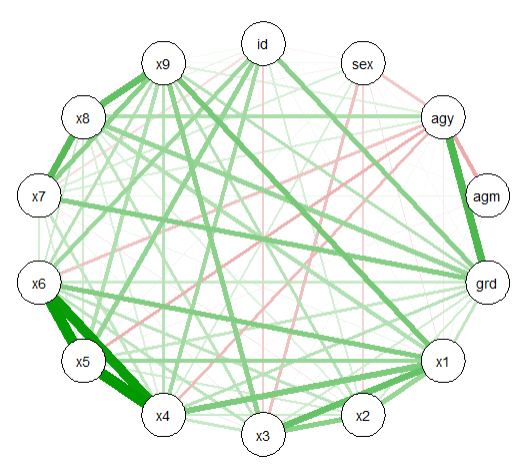
Then make the qgraph of all the correlations together. The thicker lines are stronger correlations, with green indicating positives and red for negatives. You can see in this graph for example that x4-x6 are highly correlated in the thick green triangle:
qgraph(cor_holz)
Which makes this:
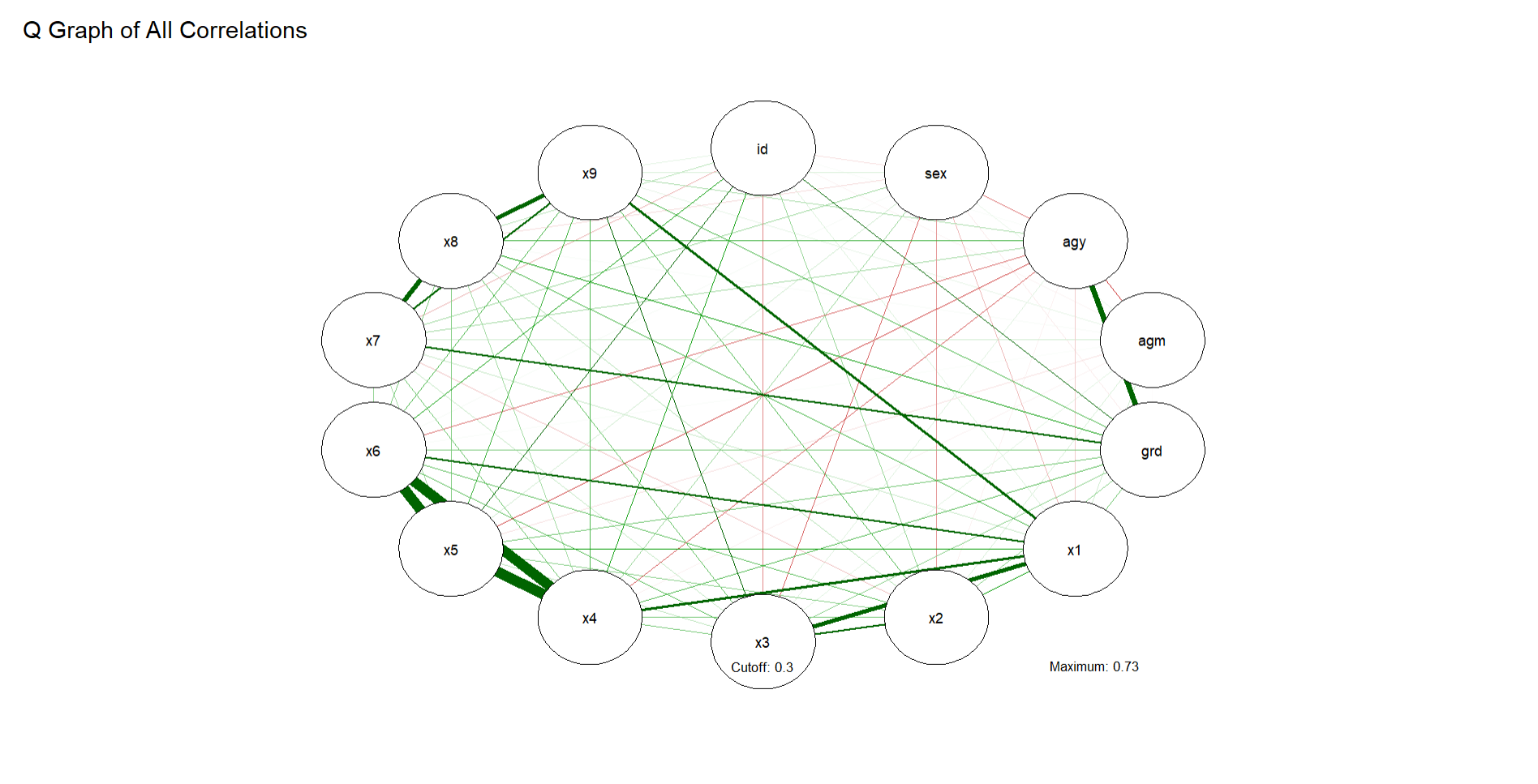
You can fancy it up a bit by establishing cutoffs for correlation values (helpful if you want to pinpoint which have the strongest correlations), add a title, and change the dimensions:
qgraph(cor_holz, # correlation
cut=.30, # cutoff value for correlations
details = T, # shows details
mar = c(6,10,6,10), # size of graph
vsize = 8, # size of nodes
title = "Q Graph of All Correlations") # title
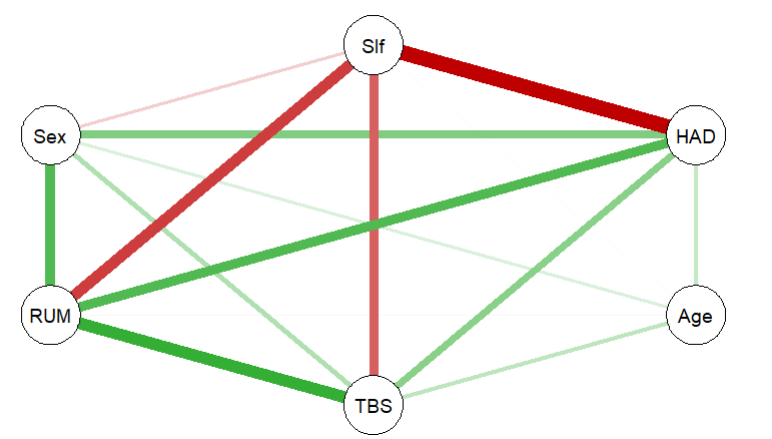
A more clear cut example is with the FacialBurns data in the same lavaan package, which shows much more obvious multicollinearity and lack thereof in the respective variables:
face_cor <- FacialBurns %>%
correlation()
qgraph(face_cor)